|
Affinity Ligands
Oligonucletides can be labeled with biotin or digoxigenin* or directly labeled with alkaline phosphatase. A variety of linker arms are available as spacers to minimize steric hindrance.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins or cognate DNA molecules by specific hybridization based affinity chromatography. The biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography.
Modifications for Oligo Synthesis
|
Product
|
Code
|
Catalog No.
|
50 nmol scale (XX=05)
|
200 nmol scale (XX=02)
|
1 umol scale (XX=01)
|
2 umol scale (XX=03)
|
5 umol scale (XX=06)
|
10 umol scale (XX=10)
|
15 umol scale (XX=15)
|
|
Biotin 3'
|
[Bio-3]
|
26-6404
|
$52.00
|
$52.00
|
$82.00
|
$127.00
|
$369.00
|
$406.00
|
$527.00
|
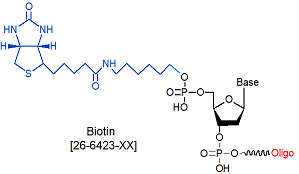
Modification : Biotin 3'
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
| Catalog No | Scale | Price | | 26-6404-05 | 50 nmol | $52.00 | | 26-6404-02 | 200 nmol | $52.00 | | 26-6404-01 | 1 umol | $82.00 | | 26-6404-03 | 2 umol | $127.00 | | 26-6404-06 | 5 umol | $369.00 | | 26-6404-10 | 10 umol | $406.00 | | 26-6404-15 | 15 umol | $527.00 |
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin 3'
|
|
Biotin deoxythymidine dT
|
[Bio-dT]
|
26-6424
|
$351.00
|
$351.00
|
$456.00
|
$684.00
|
$2,052.00
|
$3,650.00
|
$4,563.00
|
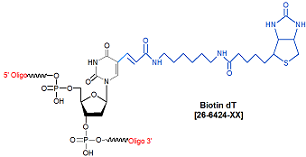
Modification : Biotin dT
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6424
Affinity Ligands
Bio-dT
Y
Y
Y
684.7
8.4
PS26-6424.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6424-05 | 50 nmol | $351.00 | | 26-6424-02 | 200 nmol | $351.00 | | 26-6424-01 | 1 umol | $456.00 | | 26-6424-03 | 2 umol | $684.00 | | 26-6424-06 | 5 umol | $2,052.00 | | 26-6424-10 | 10 umol | $3,650.00 | | 26-6424-15 | 15 umol | $4,563.00 |
| Discounts are available for Biotin dT! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin deoxythymidine dT
|
|
Biotin Multi
|
[Bio-Multi]
|
26-6417
|
$178.00
|
$178.00
|
$267.00
|
$347.00
|
$1,201.50
|
$1,064.00
|
$1,312.00
|
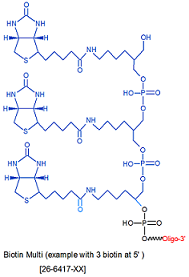
Modification : Biotin Multi
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6417
Affinity Ligands
Bio-Multi
Y
Y
Y
437.4
-
PS26-6417.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6417-05 | 50 nmol | $178.00 | | 26-6417-02 | 200 nmol | $178.00 | | 26-6417-01 | 1 umol | $267.00 | | 26-6417-03 | 2 umol | $347.00 | | 26-6417-06 | 5 umol | $1,201.50 | | 26-6417-10 | 10 umol | $1,064.00 | | 26-6417-15 | 15 umol | $1,312.00 |
| Discounts are available for Biotin Multi! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Multiple Site Note: Multi Biotin structure example shown above is for three consecutive additions. Multi Biotin sites can be added sequentially as multiple units as desired. Please indicate on your order request the number of units to be added. The code [Bio-Multi] must be added in multiples to reflect the number of sites. Example for Dual Biotin the code must be entered twice; [Bio-Multi] [Bio-Multi] and for Triple Biotin it will be [Bio-Multi] [Bio-Multi] [Bio-Multi] in the sequence where desired.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193. - Biotin Multi
|
|
Biotin NHS
|
[Bio-N]
|
26-6712
|
$210.00
|
$210.00
|
$416.00
|
$310.00
|
$546.00
|
$942.00
|
$1,253.00
|
Modification : Biotin NHS
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6712
Affinity Ligands
Bio-N
Y
Y
Y
244.32
-
PS26-6712.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6712-05 | 50 nmol | $210.00 | | 26-6712-02 | 200 nmol | $210.00 | | 26-6712-01 | 1 umol | $416.00 | | 26-6712-03 | 2 umol | $310.00 | | 26-6712-06 | 5 umol | $546.00 | | 26-6712-06 | 5 umol | $1,471.50 | | 26-6712-10 | 10 umol | $942.00 | | 26-6712-15 | 15 umol | $1,253.00 |
Click here for a list of other Affinity Ligand Modifications.
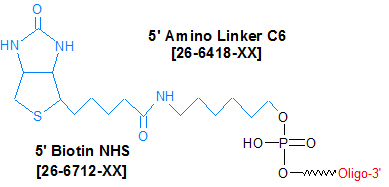
Biotin-NHS is an N-hydroxysuccinimide ester (NHS ester) of biotin. Biotin-NHS can be used to internally label an oligonucleotide with biotin at any base (that is, at a G, C, T or A position). To accomplish this, amino dG-C6/dC-C6/dA-C6/dT is first incorporated into the oligonucleotide, thereby placing an active primary amino group at the desired position. Biotin-NHS is then conjugated to the amino group in a separate reaction to form the final biotin-labeled product.
YIELD
NHS based modifications are post synthesis conjugation performed using a primary amino group. The yield is lower as compared to direct automated coupling of modifications that are available as amidites. Approximate yield for various scales are given below.
~2 nmol final yield for 50 nmol scale synthesis.
~5 nmol final yield for 200 nmol scale synthesis.
~16 nmol final yield for 1 umol scale synthesis.
~30 nmol final yield for 2 umol scale synthesis.
~75 nmol final yield for 5 umol scale synthesis.
~150 nmol final yield for 10 umol scale synthesis.
~225 nmol final yield for 15 umol scale synthesis.
Biotin is an affinity label that can be incorporated at either the 5'or 3'end of an oligonucleotide, or at an internal position. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Biotin-NHS can also be used to biotinylate a large amount of oligonucleotide aminated at the 5'or 3'end, in aqueous solution and at relatively low cost (1).
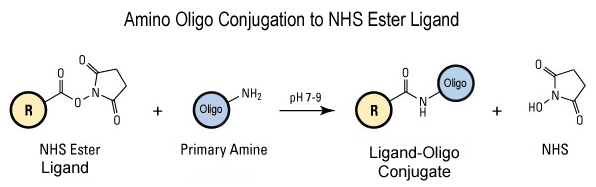
-
The primary amine labelled oligos can also be conjugated to carboxyl functional groups usually for solid supports applications using EDC mediated reaction as shown in the figure below.
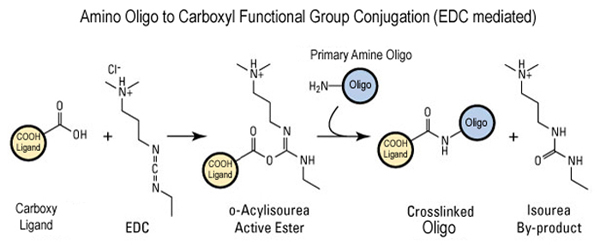
References
1. Bengtstron, M., Jungell-nortamo, A., Syvanen, A-C. Biotinylation of Oligonucleotides Using a Water Soluble Biotin Ester.Nucleos. Nucleot. Nucl. (1990), 9: 123-127.
- Biotin NHS
|
|
Biotin TEG (15 atom triethylene glycol spacer) 3'
|
[Bio-TEG-3]
|
26-6407T
|
$166.00
|
$166.00
|
$227.00
|
$272.00
|
$642.00
|
$876.00
|
$1,095.00
|
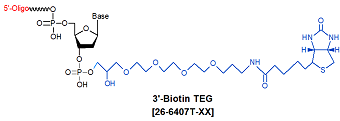
Modification : Biotin TEG 3'
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6407T
Affinity Ligands
Bio-TEG-3
Y
Y
Y
569.61
-
PS26-6407.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6407T-05 | 50 nmol | $166.00 | | 26-6407T-02 | 200 nmol | $166.00 | | 26-6407T-01 | 1 umol | $227.00 | | 26-6407T-03 | 2 umol | $272.00 | | 26-6407T-06 | 5 umol | $642.00 | | 26-6407T-10 | 10 umol | $876.00 | | 26-6407T-15 | 15 umol | $1,095.00 |
| Discounts are available for Biotin TEG 3'! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin TEG (15 atom triethylene glycol spacer) 3'
|
|
Biotin TEG (15 atom triethylene glycol spacer) 5'
|
[Bio-TEG-5]
|
26-6407F
|
$166.00
|
$166.00
|
$227.00
|
$272.00
|
$642.00
|
$876.00
|
$1,095.00
|
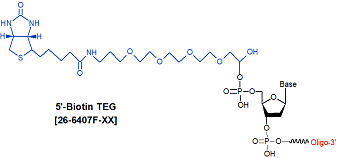
Modification : Biotin TEG 5'
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6407F
Affinity Ligands
Bio-TEG-5
Y
Y
Y
569.61
-
PS26-6407.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6407F-05 | 50 nmol | $166.00 | | 26-6407F-02 | 200 nmol | $166.00 | | 26-6407F-01 | 1 umol | $227.00 | | 26-6407F-03 | 2 umol | $272.00 | | 26-6407F-06 | 5 umol | $642.00 | | 26-6407F-10 | 10 umol | $876.00 | | 26-6407F-15 | 15 umol | $1,095.00 |
| Discounts are available for Biotin TEG 5'! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin TEG (15 atom triethylene glycol spacer) 5'
|
|
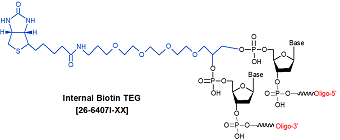
Biotin TEG (15 atom triethylene glycol spacer) Internal
|
[Bio-TEG-Int]
|
26-6407I
|
$166.00
|
$166.00
|
$227.00
|
$272.00
|
$642.00
|
$876.00
|
$1,095.00
|
Modification : Biotin TEG Internal
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6407I
Affinity Ligands
Bio-TEG-Int
Y
Y
Y
569.61
-
PS26-6407.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6407I-05 | 50 nmol | $166.00 | | 26-6407I-02 | 200 nmol | $166.00 | | 26-6407I-01 | 1 umol | $227.00 | | 26-6407I-03 | 2 umol | $272.00 | | 26-6407I-06 | 5 umol | $642.00 | | 26-6407I-10 | 10 umol | $876.00 | | 26-6407I-15 | 15 umol | $1,095.00 |
| Discounts are available for Biotin TEG Internal! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin TEG (15 atom triethylene glycol spacer) Internal
|
|
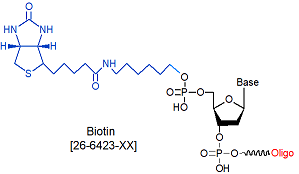
Biotin-5'
|
[Bio-5]
|
26-6423
|
$65.00
|
$65.00
|
$87.00
|
$121.00
|
$301.50
|
$442.00
|
$674.00
|
Modification : Biotin-5'
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
| Catalog No | Scale | Price | | 26-6423-05 | 50 nmol | $65.00 | | 26-6423-02 | 200 nmol | $65.00 | | 26-6423-01 | 1 umol | $87.00 | | 26-6423-03 | 2 umol | $121.00 | | 26-6423-06 | 5 umol | $301.50 | | 26-6423-10 | 10 umol | $442.00 | | 26-6423-15 | 15 umol | $674.00 |
| Discounts are available for Biotin-5'! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Biotin-5'
|
|
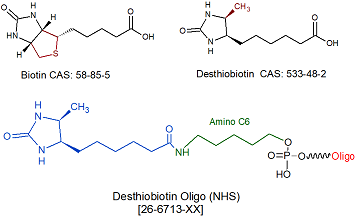
Desthiobiotin NHS
|
[DesBio-N]
|
26-6713
|
$335.00
|
$335.00
|
$459.00
|
$688.00
|
$2,065.50
|
$2,808.00
|
$3,510.00
|
Modification : Desthiobiotin NHS
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6713
Affinity Ligands
DesBio-N
Y
Y
Y
214.26
-
PS26-6713.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6713-05 | 50 nmol | $335.00 | | 26-6713-02 | 200 nmol | $335.00 | | 26-6713-01 | 1 umol | $459.00 | | 26-6713-03 | 2 umol | $688.00 | | 26-6713-06 | 5 umol | $2,065.50 | | 26-6713-10 | 10 umol | $2,808.00 | | 26-6713-15 | 15 umol | $3,510.00 |
Click here for a list of other Affinity Ligand Modifications.
Desthiobiotin modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
YIELD
NHS based modifications are post synthesis conjugation performed using a primary amino group. The yield is lower as compared to direct automated coupling of modifications that are available as amidites. Approximate yield for various scales are given below.
~2 nmol final yield for 50 nmol scale synthesis.
~5 nmol final yield for 200 nmol scale synthesis.
~16 nmol final yield for 1 umol scale synthesis
~160 nmol final yield for 10 umol scale synthesis
~240 nmol final yield for 15 umol scale synthesis
Desthiobiotin NHS & Desthiobiotin TEG
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable.
Desthiobiotin TEG is incorporated directly in an oligo and is available for 5' and 3'. Incorporation is also possible internally but it will break the nucleic acid base chain.
. Note that since Desthiobiotin NHS is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester..
Biotin
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Desthiobiotin NHS
|
|
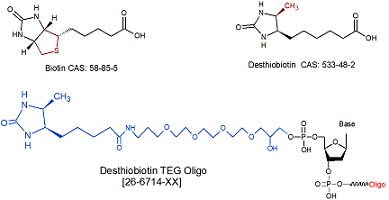
Desthiobiotin TEG
|
[DesBio-TEG]
|
26-6714
|
$443.00
|
$443.00
|
$551.00
|
$740.00
|
$2,479.50
|
$3,008.00
|
$3,548.00
|
Modification : Desthiobiotin TEG
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6714
Affinity Ligands
DesBio-TEG
Y
Y
Y
539.56
-
PS26-6714.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6714-05 | 50 nmol | $443.00 | | 26-6714-02 | 200 nmol | $443.00 | | 26-6714-01 | 1 umol | $551.00 | | 26-6714-03 | 2 umol | $740.00 | | 26-6714-06 | 5 umol | $2,479.50 | | 26-6714-10 | 10 umol | $3,008.00 | | 26-6714-15 | 15 umol | $3,548.00 |
| Discounts are available for Desthiobiotin TEG! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin TEG is incorporated directly in an oligo and is available for 5' and 3'. Incorporation is also possible internally but it will break the nucleic acid base chain.
Biotin
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193.
- Desthiobiotin TEG
|
|
Digoxigenin NHS
|
[Dig-N]
|
26-6429
|
$650.00
|
$650.00
|
$1,250.00
|
$1,875.00
|
$4,797.00
|
$6,951.00
|
$8,864.00
|
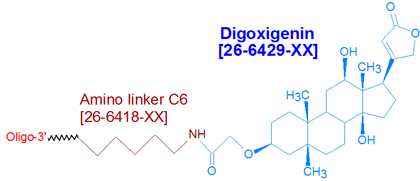
Modification : Digoxigenin NHS
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6429
Affinity Ligands
Dig-N
Y
Y
Y
561.3
13
PS26-6429.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6429-05 | 50 nmol | $650.00 | | 26-6429-02 | 200 nmol | $650.00 | | 26-6429-01 | 1 umol | $1,250.00 | | 26-6429-03 | 2 umol | $1,875.00 | | 26-6429-06 | 5 umol | $4,797.00 | | 26-6429-10 | 10 umol | $6,951.00 | | 26-6429-15 | 15 umol | $8,864.00 |
This modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C3, C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
Digoxigenin (as Digoxigenin-3-O-methylcarbonyl-epsilon-aminocaproic acid NHS ester) is a member of the steroid family found in Digitalis plants (1). It is a hapten, that is, a small molecule having high immunogenicity. Because antibodies raised against haptens have considerably higher affinities for them than other antibodies do for their targets makes haptens particularly desirable as affinity tags for oligonucleotides (2).
Digoxigenin ('Dig') is commonly used to label oligonucleotides probes for use in hybridization applications, for example, in situ hybridization (3), Northern and Southern blotting. After hybridization to their targets, these Dig-labeled probes are detected with anti-Dig antibodies that are labeled with dyes (for primary detection) or enzymes (for secondary detection using a fluorogenic, chemiluminogenic, or colorimetric substrate specific for the enzyme). To maximize signal, Gene Link recommends modifying the oligonucleotide probe with three or more Dig molecules, spaced about 10 bases apart. Note that since digoxigenin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to the digoxigenin NHS ester.
References
1. Polya, G. Biochemical targets of plant bioactive compounds. New York: CRC Press, 2003. p 847.
2. Shreder, K. Synthetic Haptens as Probes of Antibody Response and Immunorecognition. Methods (Academic Press) (2000), 20: 372-379.
3. Hauptmann, G., Gerster, T.. Two-color whole-mount in situ hybridixation to vertebrate and Drosophila embryos. Trends Genet. (1994), 10: 266.
- Digoxigenin NHS
|
|
DNP TEG (2, 4-dinitrophenyl)
|
[DNP-TEG]
|
26-6512
|
$350.00
|
$350.00
|
$456.00
|
$683.00
|
$2,052.00
|
$3,645.00
|
$4,556.00
|
.png)
Modification : DNP TEG (2, 4-dinitrophenyl)
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6512
Affinity Ligands
DNP-TEG
Y
Y
Y
509.41
1
PS26-6512.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6512-05 | 50 nmol | $350.00 | | 26-6512-02 | 200 nmol | $350.00 | | 26-6512-01 | 1 umol | $456.00 | | 26-6512-03 | 2 umol | $683.00 | | 26-6512-06 | 5 umol | $2,052.00 | | 26-6512-10 | 10 umol | $3,645.00 | | 26-6512-15 | 15 umol | $4,556.00 |
| Discounts are available for DNP TEG (2, 4-dinitrophenyl)! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
DNP (2,4-dinitrophenyl) is classified as a hapten for molecular biology purposes, that is, a small molecule having high immunogenicity. Because antibodies raised against haptens have considerably higher affinities for them than other antibodies do for their targets makes haptens particularly desirable as affinity tags for oligonucleotides (1).
DNP attached to a triethylene glycol (TEG) spacer arm is commonly used to label oligonucleotides probes for use in hybridization applications, for example, in situ hybridization, Northern and Southern blotting (2). After hybridization to their targets, these DNP-labeled probes are detected with anti-DNP antibodies that are labeled with dyes (for primary detection) or enzymes (for secondary detection using a fluorogenic, chemiluminogenic, or colorimetric (3) substrate specific for the enzyme). To maximize signal obtained with such probes, Gene Link recommends modifying the oligonucleotide probe with three DNP molecules, either grouped at the 5’-end or spaced about 10 bases apart (2).
In addition to the above straightforward anti-DNP antibody-based detection systems, oligo probes labeled with both a fluorescent dye and DNP also been used for highly-sensitive direct detection of antigens (at femtoMolar levels) in a rolling circle amplification (RCA)-based assay system (4).
References
1. Shreder, K. Synthetic Haptens as Probes of Antibody Response and Immunorecognition. Methods (Academic Press) (2000), 20: 372-379.
2. Grzybowski, J., Will, D.W., Randall, R.E., Smith, C.A., Brown, T.. Synthesis and antibody-mediated detection of oligonucleotides containing multiple 2,4-dinitrophenyl reporter groups. Nucleic Acids Res. (1993), 21: 1705-1712.
3. Lehtovaara, P., Uusi-Oukari, M., Buchert, P, Laaksonen, M., Bengtstrom, M. Ranki, M. Quantitative PCR for Hepatitis B Virus with Colorimetric Detection. Genome Res. (1993), 3: 169-175.
4. Schweitzer, B., Wiltshire, S., Lambert, J., O’Malley, S., et al. Immunoassays with rolling circle DNA amplification: A versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. (USA) (2000), 97: 10113-10119.
- DNP TEG (2, 4-dinitrophenyl)
|
|
Dual Biotin
|
[Bio-Dual]
|
26-6421
|
$350.00
|
$350.00
|
$456.00
|
$683.00
|
$2,052.00
|
$3,645.00
|
$4,556.00
|
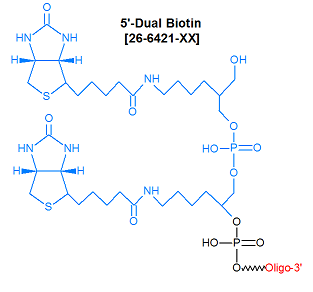
Modification : Biotin Dual
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6421
Affinity Ligands
Bio-Dual
Y
Y
N
874.8
-
PS26-6421.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6421-05 | 50 nmol | $350.00 | | 26-6421-02 | 200 nmol | $350.00 | | 26-6421-01 | 1 umol | $456.00 | | 26-6421-03 | 2 umol | $683.00 | | 26-6421-06 | 5 umol | $2,052.00 | | 26-6421-10 | 10 umol | $3,645.00 | | 26-6421-15 | 15 umol | $4,556.00 |
| Discounts are available for Biotin Dual! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Click here for a list of other Affinity Ligand Modifications.
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3) .
Desthiobiotin
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable. Note that since desthiobiotin is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester.
Desthiobiotin NHS modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
References
1. Huisgen, R. Angew. Chem. Int. Ed. (1963), 2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. (2002), 41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin. Methods Enzymol. (1970), 18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. (2002), 308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay. Appl. Environ. Microbiol. (2009), 75: 4185-4193. - Dual Biotin
|
|
NTA Mono (Nitrilotriacetate) Oligo Conjugate
|
[NTA]
|
26-6444
|
$478.00
|
$478.00
|
$675.00
|
$1,012.00
|
$3,037.50
|
$4,752.00
|
$5,702.00
|
Modification : NTA Mono
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
| Catalog No | Scale | Price | | 26-6444-05 | 50 nmol | $478.00 | | 26-6444-02 | 200 nmol | $478.00 | | 26-6444-01 | 1 umol | $675.00 | | 26-6444-03 | 2 umol | $1,012.00 | | 26-6444-06 | 5 umol | $3,037.50 | | 26-6444-10 | 10 umol | $4,752.00 | | 26-6444-15 | 15 umol | $5,702.00 |
NTA-Oligo modified with nitrilotriacetate (NTA)is a post synthesis conjugation to thiol. The thiol group can be placed at the 5' and 3' and for internal positions DTSPA can be used. The price listed is for NTA modification and does not include thiol modification, oligo synthesis and purification charges
Nitrilotriacetic acid (NTA) is widely used in affinity chromatography to purify His-tagged proteins.
The NTA chelator forms a complex with metal ions, usually Ni(2+), which then binds to histidine residues in the His-tag.
Two forms of NTA are available, monovalent (NTA Mono) and trivalent (NTA Tris) differ in their binding strength and application.
Tris-NTA has a significantly higher affinity for His-tags compared to mono-NTA, with affinities in the nanomolar range versus the micromolar range for mono-NTA.
This higher affinity is due to the multivalency of tris-NTA, which allows it to bind the His-tag at three points, resulting in a much more stable and strong interaction.
This leads to more stable baselines in experiments like SPR and improved detection sensitivity.
Comparison of NTA Mono and NTA Tris Affinity & Other Features Towards His Tag
|
Feature
|
Mono NTA
|
Tris NTA
|
| Structure |
NTA Mono: One NTA group per ligand |
NTA Tris: Three NTA groups per ligand (clustered) |
| Histidine Binding |
Binds via 2-4 histidine residues |
Binds via 6+ residues, higher avidity |
| Affinity |
uM range K_D |
nM-pM range K_D |
| Elution |
Easy with imidazole/EDTA |
Difficult due to strong binding.
Requires 100 mM imidazole or higher |
| Application |
Protein purification. Weak binding acceptable |
Requiring high affinity binding.
Biosensing, stable immobilization |
|
The enhanced binding of tris-NTA is a result of multivalency, where the simultaneous binding of three NTA-metal ion complexes to the histidine tag creates a much stronger overall interaction.
|
Applications
- NTA Mono is commonly used in immobilized metal affinity chromatography (IMAC) for routine purification of His-tagged proteins.
- NTA Tris provides higher binding strength and is preferred in biosensor applications such as SPR (Surface Plasmon Resonance) and BLI (Bio-Layer Interferometry).
Binding Affinity Summary
|
Ligand
|
Approximate K_D to His tag
|
| NTA Mono-Ni(2+) |
~1-10 uM |
| NTA Tris-Ni(2+) |
~1-100 nM |
|
The enhanced binding of tris-NTA is a result of multivalency, where the simultaneous binding of three NTA-metal ion complexes to the histidine tag creates a much stronger overall interaction.
The His-tagged protein is effectively "gripped" by three separate NTA-metal interactions. Dissociation requires breaking all three bonds simultaneously, which is statistically far less likely and requires much more energy.
|
References
1. 1. J. Shimada, T. Maruyama, T. Hosogi, J. Tominaga, N. Kamiya, M. Goto,. Conjugation of DNA with protein using His-tag chemistry and its application to the aptamer-based detection system, Biotechnol. Lett. 30 (2008) 2001-2006.
2.J. Shimada et al DNA enzyme conjugate that can detect thrombin. Anal. Biochem. 414 (2011) 103-108.
- NTA Mono (Nitrilotriacetate) Oligo Conjugate
3. Hochuli, E., et al. (1988). Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. *Bio/Technology*, 6, 1321-1325
4. Lata, S., et al. (2005). High-affinity adaptors for switchable recognition of histidine-tagged proteins. *Journal of the American Chemical Society*, 127(29), 10205-10215.
5. Porath, J., et al. (1975). Metal chelate affinity chromatography, a new approach to protein fractionation. *Nature*, 258, 598-599.
- NTA Mono (Nitrilotriacetate) Oligo Conjugate
|
|
Oligo Affinity Support Non Cleavable
|
[OAS]
|
26-6510
|
$245.00
|
$245.00
|
$310.00
|
$410.00
|
$625.00
|
$880.00
|
$1,450.00
|
Modification : Oligo Affinity Support
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
| Catalog No | Scale | Price | | 26-6510-05 | 50 nmol | $245.00 | | 26-6510-02 | 200 nmol | $245.00 | | 26-6510-01 | 1 umol | $310.00 | | 26-6510-03 | 2 umol | $410.00 | | 26-6510-06 | 5 umol | $625.00 | | 26-6510-10 | 10 umol | $880.00 | | 26-6510-15 | 15 umol | $1,450.00 |
Oligo Affinity Support. Non-Cleavable Matrix.
Gene Link also offers oligos covalently linked to magnetic beads to be used as affinity support for the purification, capture and elution of the complimentary sequences. Oligo Magnetic Beads.
Oligo Affinity Support is a non-cleavable support that can also be used for the purification, capture and elution of the complimentary sequences.
Researchers are able to choose from a wide selection of techniques for attaching oligonucleotides to solid supports. However, these techniques usually require synthesis and purification of an amino-modified oligonucleotide, followed by its covalent linkage to an appropriately activated solid support. While these techniques are relatively straightforward, there still exists a need to prepare affinity supports simply and directly.
The Oligo-Affinity Support (OAS)5 consists of fibers with an inner core of Teflon, covalently coated with an organic layer of functionalized copolymers. A 25-carbon spacer arm terminating in adenosine with a DMT group at the 5'-position is attached to the support. Since the linkage to adenosine is stable to acid and base, the oligonucleotide formed by the standard synthesis protocol remains attached to the support after deprotection.
For use as an affinity support for the purification of DNA binding proteins6, one oligonucleotide was synthesized and deprotected using ammonium hydroxide on the support. The complementary strand was then bound through normal hybridization. The affinity support was used to purify a sequence specific DNA binding protein 100 fold to near homogeneity. The Teflon support showed very low levels of non-specific binding of proteins. It also exhibited the further advantage of being composed of non-friable flexible fibers which do not shrink or swell. The matrix proved to be stable and reusable at least ten times.
Although the most likely use for the OAS is in the preparation of affinity supports, ligations and kinase reactions can also be carried out.
References
1. Ruth, J., Smith, R.D., Lohrmann, R. Fed. Proc. 1985, 44, 1622.
2. Duncan, C.H. and Cavalier,S.L. Analytical Biochemistry, 1988, 169, 104.
3. . Kadonaga, J.T Methods Enzymol, 1991, 208, 10-23.
4. Kirch, H.C.; Kruger,H and H.S. Holthausen, Nucleic Acids Res., 1991, 19, 3156.
5. Larson, C.L. and Verdine,G.L. Nucleic Acids Res., 1992, 20, 3525.
- Oligo Affinity Support Non Cleavable
|
|
pseudoUridine ribo (psi rU)
|
[psi-rU]
|
27-6531
|
$643.00
|
$643.00
|
$751.00
|
$845.00
|
$3,379.50
|
$5,227.00
|
$6,426.00
|
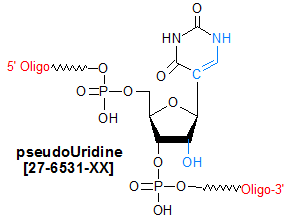
Modification : pseudoUridine (psi rU)
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
27-6531
Affinity Ligands
psi-rU
Y
Y
Y
306.17
9.9
PS27-6531.pdf
-
-
-
| Catalog No | Scale | Price | | 27-6531-05 | 50 nmol | $643.00 | | 27-6531-02 | 200 nmol | $643.00 | | 27-6531-01 | 1 umol | $751.00 | | 27-6531-03 | 2 umol | $845.00 | | 27-6531-06 | 5 umol | $3,379.50 | | 27-6531-10 | 10 umol | $5,227.00 | | 27-6531-15 | 15 umol | $6,426.00 |
| Discounts are available for pseudoUridine (psi rU)! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Pseudouridine-("psi") is a C-glyoside isomer of uridine, and is the most common modified nucleoside found in structural RNA, such as tRNA, rRNA, snRNA, and snoRNA (1,2). Psi-modified RNA can be used as research tools for studies into the roles of this residue in RNA structure and function in the cell. Currently, the role of psi in RNA is a subject of active research, with some things now known. Psi can coordinate a water molecule through its free N1 hydrogen, thereby inducing a modest increase in rigidity on the nearby sugar-phosphate backbone. The presence of psi also enhances base-stacking. Such effects have been proposed as explanations for the deleterious functional effects observed in mutant bacterial strains that lack certain psi residues in tRNA or rRNA (2). Also, based on recent studies, it has been proposed that psi may offer RNA molecules protection from radiation (3).
References
1. Hamma, T., Ferre-D-Amare, A.R. Pseudouridine synthases. Chem. Biol. (2006), 13: 1125-1135.
2. Charette, M., Gray, M.W. Pseudouridine in RNA: what, where, how, and wny. IUBMB Life (2000), 49: 341-351.
3. Monobe, M., Arimoto-Kobayashi, S., Ando, K. beta-Pseudouridine, a beer component, reduces radiation-induced chromosome aberrations in human lymphocytes. Mut. Res-Genet. Toxicol. and Env. Mutagen. (2003), 538: 93-99.
- pseudoUridine ribo (psi rU)
|
|
Stearyl C18 LMO LMO 3'
|
[Str-C18-3]
|
26-6623
|
$200.00
|
$200.00
|
$255.00
|
$362.00
|
$1,147.50
|
$1,782.00
|
$2,144.00
|
-3'-LMO.png)
Modification : Stearyl C18 (3') LMO
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6623
Affinity Ligands
Str-C18-3
N
Y
N
460.61
-
PS26-6623.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6623-05 | 50 nmol | $200.00 | | 26-6623-02 | 200 nmol | $200.00 | | 26-6623-01 | 1 umol | $255.00 | | 26-6623-03 | 2 umol | $362.00 | | 26-6623-06 | 5 umol | $1,147.50 | | 26-6623-10 | 10 umol | $1,782.00 | | 26-6623-15 | 15 umol | $2,144.00 |
Oligonucleotides are predominantly hydrophilic species and require help in permeating cell membranes. One strategy to improve cellular uptake of therapeutic oligonucleotides is to conjugate them with non-toxic, lipophilic molecules. Gene Link offers cholesteryl TEG, alpha-tocopherol and stearyl labelling of oligonucleotides and this strategy has proved to be useful for delivering therapeutic oligonucleotides to a broad distribution of targets.
Stearyl Modification
Stearyl Modification is C18 lipid, it is an economical and effective carrier molecule. We envisage that the 5'-stearyl group will become a favored lipophilic carrier for experimentation with synthetic oligonucleotides.
Cholesterol TEG Modification
Cholesterol TEG Modification is a lipophilic modification aiding in cellular delivery. The TEG liker arm facilitates solubility issues of the oligo making it soluble in aqueous buffers.
alpha-tocopherol TEG Modification
Similar to cholesterol TEG, alpha-tocopherol (vitamin E) is both lipophilic and non-toxic even at high doses so would be an excellent candidate as a lipophilic carrier for oligonucleotides. The TEG liker arm facilitates solubility issues of the oligo making it soluble in aqueous buffers.
GalNAc
A more directed approach to the delivery of therapeutic oligonucleotides specifically to the liver has been to target the asialoglycoprotein receptor (ASGPR) using a suitable glycoconjugate. Indeed, ASGPR is the ideal target for delivery of therapeutic oligonucleotides to the liver since it combines tissue specificity, high expression levels and rapid internalization and turnover. The use of oligonucleotide glycoconjugates has led to significant advances in therapeutic delivery as evidenced by the work of Alnylam Pharmaceuticals which has developed multivalent N-acetylgalactosamine (GalNAc) conjugated siRNAs that bind at nanomolar levels to ASGPR (1). A similar strategy has been applied at Ionis Pharmaceuticals directed at the development of antisense oligonucleotide therapeutics (2).
The GalNAc ligand originally used by Alnylam is the triantennary ligand would seem to lend itself to formation by post synthesis conjugation to the 3' terminus but a complex trivalent GalNAc support would also be perfectly applicable, if challenging to produce. However, an alternative approach using a monovalent GalNAc support with two additions of a monovalent GalNAc phosphoramidite was also described and yielded a trivalent GalNAc structure. This (1+1+1) trivalent GalNAc structure led to GalNAc modified siRNA oligos with potency equal to the equivalent siRNA with the triantennary GalNAc ligand both in vitro and in vivo.
Researchers at Ionis have developed antisense oligonucleotides containing the GalNAc cluster. In their case, they were able to show2 that moving the triantennary GalNAc ligand to the 5' terminus led to improved potency in vitro and in vivo. As may be expected, such a large complex ligand lends itself to solution phase chemistry to produce GalNAc modified antisense oligos. However, a solid phase synthetic approach was also described, and compared to the solution phase approach structure of the 5'-GalNAc triantennary ligand (4).
A further report on antisense oligonucleotides demonstrated (5) the effectiveness of modifying at the 5' terminus using monovalent GalNAc ligands. Up to five GalNAc monomers were added in a serial manner (Figure 3) and it was shown that activity of the antisense oligonucleotides improved as the number of GalNAc units increased. The authors also showed that phosphodiester linkages between the GalNAc units were preferable to phosphorothioate linkages in their testing (5).
Recommended Further Reading
N-acetylgalactosamine (GalNAc) Oligo Application Note: Glen Report 29.14: N-acetylgalactosamine (GalNAc) Oligonucleotide Conjugates
References. Adapted from Glen Research Reports. http://www.glenresearch.com/GlenReports/GR29-14.html
1. J.K. Nair, et al., J Am Chem Soc, 2014, 136, 16958-61.
2. T.P. Prakash, et al., Bioorganic & Medicinal Chemistry Letters, 2015, 25, 4127-4130.
3. K.G. Rajeev, et al., Chembiochem, 2015, 16, 903-8.
4. I. Cedillo, et al., Molecules, 2017, 22.
5. T. Yamamoto, M. Sawamura, F. Wada, M. Harada-Shiba, and S. Obika, Bioorganic & Medicinal Chemistry, 2016, 24, 26-32.
- Stearyl C18 LMO LMO 3'
|
|
Tris NTA (Tris Nitrilotriacetate) Oligo Conjugate
|
[NTA-Tris]
|
26-6614
|
$985.00
|
$985.00
|
$1,380.00
|
$1,680.00
|
$2,240.00
|
$4,480.00
|
$7,185.00
|
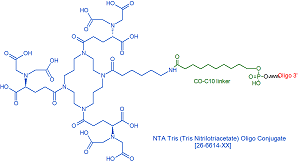
Modification : NTA Tris
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6614
Affinity Ligands
NTA-Tris
Y
N
N
1049
-
PS26-6614.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6614-05 | 50 nmol | $985.00 | | 26-6614-02 | 200 nmol | $985.00 | | 26-6614-01 | 1 umol | $1,380.00 | | 26-6614-03 | 2 umol | $1,680.00 | | 26-6614-06 | 5 umol | $2,240.00 | | 26-6614-10 | 10 umol | $4,480.00 | | 26-6614-15 | 15 umol | $7,185.00 |
Gene Link provides custom synthesis of NTA Tris (Nitrilotriacetate) Oligo Conjugate.
NTA-Tris modification is a post synthesis conjugation to an active NHS group of either a NHS-Carboxy C10 or a NHS-dT group thus an additional modification is required at the 5' end with additional charges for that modification.
Nitrilotriacetic acid (NTA) is widely used in affinity chromatography to purify His-tagged proteins.
The NTA chelator forms a complex with metal ions, usually Ni(2+), which then binds to histidine residues in the His-tag.
Two forms of NTA are available, monovalent (NTA Mono) and trivalent (NTA Tris) differ in their binding strength and application.
Tris-NTA has a significantly higher affinity for His-tags compared to mono-NTA, with affinities in the nanomolar range versus the micromolar range for mono-NTA.
This higher affinity is due to the multivalency of tris-NTA, which allows it to bind the His-tag at three points, resulting in a much more stable and strong interaction.
This leads to more stable baselines in experiments like SPR and improved detection sensitivity.
Comparison of NTA Mono and NTA Tris Affinity & Other Features Towards His Tag
|
Feature
|
Mono NTA
|
Tris NTA
|
| Structure |
NTA Mono: One NTA group per ligand |
NTA Tris: Three NTA groups per ligand (clustered) |
| Histidine Binding |
Binds via 2-4 histidine residues |
Binds via 6+ residues, higher avidity |
| Affinity |
uM range K_D |
nM-pM range K_D |
| Elution |
Easy with imidazole/EDTA |
Difficult due to strong binding.
Requires 100 mM imidazole or higher |
| Application |
Protein purification. Weak binding acceptable |
Requiring high affinity binding.
Biosensing, stable immobilization |
|
The enhanced binding of tris-NTA is a result of multivalency, where the simultaneous binding of three NTA-metal ion complexes to the histidine tag creates a much stronger overall interaction.
|
Applications
- NTA Mono is commonly used in immobilized metal affinity chromatography (IMAC) for routine purification of His-tagged proteins.
- NTA Tris provides higher binding strength and is preferred in biosensor applications such as SPR (Surface Plasmon Resonance) and BLI (Bio-Layer Interferometry).
Binding Affinity Summary
|
Ligand
|
Approximate K_D to His tag
|
| NTA Mono-Ni(2+) |
~1-10 uM |
| NTA Tris-Ni(2+) |
~1-100 nM |
|
The enhanced binding of tris-NTA is a result of multivalency, where the simultaneous binding of three NTA-metal ion complexes to the histidine tag creates a much stronger overall interaction.
The His-tagged protein is effectively "gripped" by three separate NTA-metal interactions. Dissociation requires breaking all three bonds simultaneously, which is statistically far less likely and requires much more energy.
|
References
1. 1. J. Shimada, T. Maruyama, T. Hosogi, J. Tominaga, N. Kamiya, M. Goto,. Conjugation of DNA with protein using His-tag chemistry and its application to the aptamer-based detection system, Biotechnol. Lett. 30 (2008) 2001-2006.
2.J. Shimada et al DNA enzyme conjugate that can detect thrombin. Anal. Biochem. 414 (2011) 103-108.
- NTA Mono (Nitrilotriacetate) Oligo Conjugate
3. Hochuli, E., et al. (1988). Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. *Bio/Technology*, 6, 1321-1325
4. Lata, S., et al. (2005). High-affinity adaptors for switchable recognition of histidine-tagged proteins. *Journal of the American Chemical Society*, 127(29), 10205-10215.
5. Porath, J., et al. (1975). Metal chelate affinity chromatography, a new approach to protein fractionation. *Nature*, 258, 598-599.
- Tris NTA (Tris Nitrilotriacetate) Oligo Conjugate
|
|
Tyramide C10 Oligo-5'
|
[Tyr-C10]
|
26-6681
|
$330.00
|
$330.00
|
$448.00
|
$578.00
|
$885.00
|
$1,663.00
|
$1,995.00
|
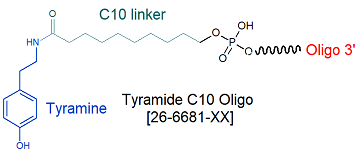
Modification : Tyramide C10 Oligo-5'
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
| Catalog No | Scale | Price | | 26-6681-05 | 50 nmol | $330.00 | | 26-6681-02 | 200 nmol | $330.00 | | 26-6681-01 | 1 umol | $448.00 | | 26-6681-03 | 2 umol | $578.00 | | 26-6681-06 | 5 umol | $885.00 | | 26-6681-10 | 10 umol | $1,663.00 | | 26-6681-15 | 15 umol | $1,995.00 |
| Discounts are available for Tyramide C10 Oligo-5'! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Tyramide-conjugated DNA barcodes & Tyramide Signal Amplification
See a report by Paul Simonson, Itzel Valencia, Sanjay S. Patel. Indexable signal amplification for multiparametric imaging.
in bioRxiv preprint.
Multiparametric imaging allows researchers to measure the expression of many biomarkers simultaneously, allowing detailed characterization of cell microenvironments. One such technique, CODEX, allows fluorescence imaging of >30 proteins in a single tissue section. In the commercial CODEX system, primary antibodies are conjugated to DNA barcodes. This modification can result in antibody dysfunction, and development of a custom antibody panel can be very costly and time consuming as trial and error of modified antibodies proceeds. To address these challenges, we developed novel tyramide-conjugated DNA barcodes that can be used with primary antibodies via peroxidase-conjugated secondary antibodies. This approach results in signal amplification and imaging without the need to conjugate primary antibodies. When combined with commercially available barcode-conjugated primary antibodies, working antibody panels can be quickly developed. This report presents methods to perform antibody staining using a commercially available automated tissue stainer and in situ hybridization imaging on a CODEX platform.
In this report, they authors present a generalizable method for CODEX imaging with unmodified primary antibodies and signal amplification that uses novel tyramide-conjugated DNA barcodes (tyramide-barcodes). This allows the benefits of tyramide signal amplification, but image the targets in an indexed way using a commercial CODEX imaging system. This approach is flexible, and demonstrate that it can be used with in situ hybridization probes. In order to make this system workable, they developed a staining approach that allowed to perform both tyramide-barcode staining and standard CODEX staining using a commercially available automated tissue stainer, made possible using a custom coverslip holder. This drastically reduces the amount of manual labor required for CODEX staining and has the potential to improve staining reproducibility.
This modification is a post synthesis conjugation to an oligo with an active NHS group of Carboxy C10 modification.
- Tyramide C10 Oligo-5'
|
|
Tyramide CO dT oligo Internal
|
[Tyr-CO-dT]
|
26-6523
|
$490.00
|
$490.00
|
$585.00
|
$645.00
|
$1,210.00
|
$1,865.00
|
$2,375.00
|
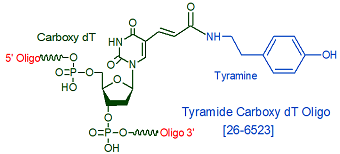
Modification : Tyramide CO dT oligo
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6523
Affinity Ligands
Tyr-CO-dT
Y
N
N
480.4
-
PS26-6523.pdf
-
-
-
| Catalog No | Scale | Price | | 26-6523-05 | 50 nmol | $490.00 | | 26-6523-02 | 200 nmol | $490.00 | | 26-6523-01 | 1 umol | $585.00 | | 26-6523-03 | 2 umol | $645.00 | | 26-6523-06 | 5 umol | $1,210.00 | | 26-6523-10 | 10 umol | $1,865.00 | | 26-6523-15 | 15 umol | $2,375.00 |
| Discounts are available for Tyramide CO dT oligo! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Tyramide-conjugated DNA barcodes & Tyramide Signal Amplification
See a report by Paul Simonson, Itzel Valencia, Sanjay S. Patel. Indexable signal amplification for multiparametric imaging.
in bioRxiv preprint.
Multiparametric imaging allows researchers to measure the expression of many biomarkers simultaneously, allowing detailed characterization of cell microenvironments. One such technique, CODEX, allows fluorescence imaging of >30 proteins in a single tissue section. In the commercial CODEX system, primary antibodies are conjugated to DNA barcodes. This modification can result in antibody dysfunction, and development of a custom antibody panel can be very costly and time consuming as trial and error of modified antibodies proceeds. To address these challenges, we developed novel tyramide-conjugated DNA barcodes that can be used with primary antibodies via peroxidase-conjugated secondary antibodies. This approach results in signal amplification and imaging without the need to conjugate primary antibodies. When combined with commercially available barcode-conjugated primary antibodies, working antibody panels can be quickly developed. This report presents methods to perform antibody staining using a commercially available automated tissue stainer and in situ hybridization imaging on a CODEX platform.
In this report, they authors present a generalizable method for CODEX imaging with unmodified primary antibodies and signal amplification that uses novel tyramide-conjugated DNA barcodes (tyramide-barcodes). This allows the benefits of tyramide signal amplification, but image the targets in an indexed way using a commercial CODEX imaging system. This approach is flexible, and demonstrate that it can be used with in situ hybridization probes. In order to make this system workable, they developed a staining approach that allowed to perform both tyramide-barcode staining and standard CODEX staining using a commercially available automated tissue stainer, made possible using a custom coverslip holder. This drastically reduces the amount of manual labor required for CODEX staining and has the potential to improve staining reproducibility.
This modification is a post synthesis conjugation to an oligo with an active NHS group of Carboxy C10 modification.
- Tyramide CO dT oligo Internal
|
|


