Modification : Desthiobiotin NHS
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6713
Affinity Ligands
[DesBio-N]
Y
Y
Y
214.26
-
PS26-6713.pdf
-
-
-
| Catalog No | Scale | Price |
| 26-6713-05 | 50 nmol | $335.00 |
| 26-6713-02 | 200 nmol | $335.00 |
| 26-6713-01 | 1 umol | $459.00 |
| 26-6713-03 | 2 umol | $688.00 |
| 26-6713-06 | 5 umol | $2,065.50 |
| 26-6713-10 | 10 umol | $2,808.00 |
| 26-6713-15 | 15 umol | $3,510.00 |
Click here for a list of other Affinity Ligand Modifications.
Desthiobiotin modification is a post synthesis conjugation to a primary amino group thus an additional modification with an amino group is required. A C6 or C12 amino group can be placed at the 5' or for the 3' end a C3 or C7 amino and for internal positions an amino modified base is used, e.g Amino dT C6.
YIELD
NHS based modifications are post synthesis conjugation performed using a primary amino group. The yield is lower as compared to direct automated coupling of modifications that are available as amidites. Approximate yield for various scales are given below.
~2 nmol final yield for 50 nmol scale synthesis.
~5 nmol final yield for 200 nmol scale synthesis.
~16 nmol final yield for 1 umol scale synthesis
~160 nmol final yield for 10 umol scale synthesis
~240 nmol final yield for 15 umol scale synthesis
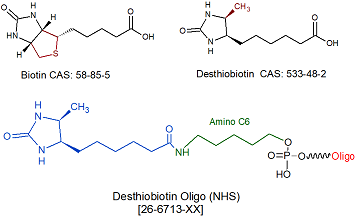
Desthiobiotin NHS & Desthiobiotin TEG
Desthiobiotin is a biotin derivative. Like biotin, desthiobiotin binds to streptavidin, but its binding affinity is considerably less (2x10E-9 M) than that of biotin (4.0x10E-14 M) (1). Consequently, oligonucleotides labeled with desthiobiotin can be easily displaced from streptavidin by biotin, thereby making recovery of the labeled oligo (for example, in affinity purification protocols) from a streptavidin-coated support a relatively simple process (2). Desthiobiotin-labeled oligos can also be conveniently eluted from streptavidin-coated supports by incubation in distilled water at 95C for 10 minutes (3). Gene Link recommends substitution of of desthiobiotin for biotin for those cases where reversible capture of oligonucleotides is desirable.
Desthiobiotin TEG is incorporated directly in an oligo and is available for 5' and 3'. Incorporation is also possible internally but it will break the nucleic acid base chain.
. Note that since Desthiobiotin NHS is in the form of an NHS ester, an active primary amino group (such as Amino Linker C6) must first be incorporated into the oligonucleotide, to allow for subsequent conjugation to desthiobiotin NHS ester..
Biotin
Biotin is an affinity label that can be incorporated at either the 5'- or 3'-end of an oligonucleotide, or at an internal position using biotin dT or Amino bases for conjugation to biotin-NHS. Biotin has a high affinity for the bacterial protein, streptavidin, which can be conjugated to a solid support (such as magnetic beads) for use as a capture and immobilization medium for a biotinylated oligo. In the biotin phosphoramidite, the biotin is attached to a long spacer arm, which acts to minimize steric hindrance between the biotin moiety and the oligo, thereby providing streptavidin easy access to the biotin. Biotinylated oligos are most commonly used as probes or primers in a variety of in vitro and in vivo applications.
Besides their importance as nucleic acid probes, biotinylated oligonucleotides are also useful for the purification of DNA binding proteins. In this context, the biotinylated oligonucleotide can be bound to a streptavidin matrix and used for either column or spin chromatography. For isolation of DNA binding proteins, the streptavidin-biotin-oligonucleotide complex is incubated with a crude cell extract containing nuclear proteins. Following appropriate washes, the proteins that bind selectively to the oligonucleotide sequence can be eluted under conditions that disrupt the protein:DNA complex. Because the binding of biotin to streptavidin is essentially irreversible and is resistant to chaotropic agents and extremes of pH and ionic strength, the elution conditions can be relatively stringent.
Dual Biotin
Dual Biotin modification specifically can be used to add multiple biotin moieties at the 5'- or 3'-end of an oligo. The most common use of this modification is to incorporate two biotin (Dual Biotin) molecules in sequence (separated by a six-carbon linker) at the 5'-end of an oligo. This "Dual Biotin" has higher binding affinity for streptavidin than that of a single biotin. The additional binding strength can be critical for applications requiring the use of biotinylated DNA attached to streptavidin-coated beads at higher temperature (for example, in PCR). Dual Biotin is known to prevent or effectively reduce loss of biotinylated DNA from such beads during heating (1). Dual biotin also is used to label the linker primers in Serial Analysis of Gene Expression (SAGE) protocols (2).
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3)
.
Multi Biotin
Multi-Biotin is a modification that can be added sequentially as many units as desired based on the application. Use Dual Biotin modification code [Bio-Dual] that is specific for two units of biotin.
For direct biotin-labeling of target RNA transcripts for microarray analysis, a special 3'-biotinylated donor nucleotide molecule containing three biotin molecules in sequence was synthesized and then ligated to the target RNA using T4 RNA ligase. The attachment of three biotins to RNA in this manner resulted in a 30% increase in target signal intensity and improved transcript detection sensitivity (3)
.
References
1. Huisgen, R.
Angew. Chem. Int. Ed. (1963),
2: 565-568.
2. Rostovtsev, V.V., Green, L.G., Fokin, V.V., Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes.
Angew. Chem. Int. Ed. (2002),
41: 2596-2599.
3. Green, N.M. Spectrophotometric determination of avidin and biotin.
Methods Enzymol. (1970),
18A: 418-424.
4. Hirsch, J.D., Eslamizar, L., Filanoski, B.J., Malekzadeh, N., Haugland, R.P., Beechem, J.M., Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation.
Anal. Biochem. (2002),
308: 343-357.
5. van Doom, R., Slawiak, M., Szemes, M., Dullemans, A.M., Bonants, P., Kowalchuk, G.A., Schoen, C.D. Robust Definition and Identification of Multiple Oomycetes and Fungi in Environmental Samples by Using a Novel Cleavable Padlock Probe-Based Ligation Detection Assay.
Appl. Environ. Microbiol. (2009),
75: 4185-4193.
- Desthiobiotin NHS
