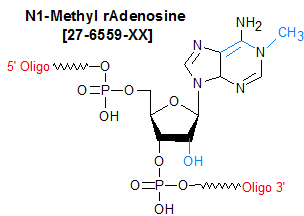
Modification : N1-Methyl rA (m1A)
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
27-6559
Structural Studies
[m1A]
Y
Y
Y
344.24
15.4
PS27-6559.pdf
-
-
-
| Catalog No | Scale | Price |
| 27-6559-05 | 50 nmol | $585.00 |
| 27-6559-02 | 200 nmol | $585.00 |
| 27-6559-01 | 1 umol | $634.00 |
| 27-6559-03 | 2 umol | $800.00 |
| 27-6559-06 | 5 umol | $2,403.00 |
| 27-6559-10 | 10 umol | $4,268.00 |
| 27-6559-15 | 15 umol | $5,335.00 |
| Discounts are available for N1-Methyl rA (m1A)! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Dimroth rearrangement of m1A to m6A under basic conditions.
Our Synthesis and deprotection scheme of mild conditions for deprotection of synthesized oligos for 60 hours at room temperature usually has no rearrangement.
Methylation of adenosine at position 1 produces a drastic functional change in the nucleobase. 1-Methyladenosine (pKa 8.25) is a much stronger base than adenosine (pKa 3.5). N-1 methylation excludes participation of the adenine base in canonical Watson-Crick base pairing and provides a positive charge to the nucleobase. This modification also alters the hydrophobicity of the base, the stacking properties, the ordering of water molecules and the chelation properties. The base may become involved in non-canonical hydrogen bonding, in electrostatic interactions and, in general, it may contribute to the conformational dynamics of the tRNA.
N1-Methyl riboadenosine (N1-Me-rA; also known as 1-Me-rA) is a methylated RNA nucleoside base, and is primarily used in the study of its role in tRNA folding. N1-Me-rA occurs in nature as a post-transcriptional modification, in which the N1 position of adenine is methylated by methyl-1-adenosine transferase (1). In tRNA, N1-Me-rA often is found at position 58 in the T loop, and position 14 in the D loop (2). Its presence introduces significant 3-D structural alteration to the tRNA (3); these alterations can be necessary for establishment of reverse transcription in virus-infected cells. For example, in order for HIV-1 to successfully infect cells, it must be able to divert tRNALys in the cell for reverse transcription; in order to do this, position 58 of the tRNA must be N3-Me-rA (4).
References
(1) Anderson, J., Droogmans, L. Biosynthesis and function of 1-Me-Ado in tRNA. In
Topics in Current Genetics, Springer Eds., 2005, 12: 121-139.
(2) Sierzputowska-Gracz, H., Gopal, D., Agris, P.F. Comparative structural analysis of 1-methyladenosine, 7-methylguanosine, ethenodenosine, and their protonated salts. IV. 1H, 13C and 15N NMR studies at natural isotope abundance.
Nucleic Acids Res. (1986),
14: 7783-7801.
(3) Helm, M., et al. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA.
Nucleic Acids Res. (1998),
26: 1636-1643.
(4) Marquet, R., Dardel, F. Transfer RNA modifications and DNA editing in HIV-1 reverse transcription. In
Topics in Current Genetics, Springer Eds., 2005, 12: 401-429.
- N1-Methyl rAdenosine (m1A)
