| Home | Products | Tools | Technical Resource | Catalog Order | Custom Oligo Order |
 |
|
Technical Sheet q Catalog No. 40-2001-10 10 nmoles
Shipped at ambient temperature. Store at -20oC |
||||||||||||||||
|
Background
Sickle cell anemia is an autosomal recessive disease. A single base change (A to T) in the b globin chain causes the substitution of amino acid glutamine to valine; the cause of the disorder sickle cell anemia. The resulting mutant globin chain is termed as the Hb S. Hemoglobin S is freely soluble when fully oxygenated, under conditions of low oxygen tension the red cells become grossly abnormal assuming a sickle shape leading to aggregation and hemolysis. Homozygous Hb S is a serious hemoglobinopathy found almost exclusively in the Black population. About 8% of American Blacks are carriers and about 0.2% are affected. Heterozygotes (sickle cell trait) are clinically normal, although their red cells will sickle when subjected to very low oxygen pressure in vitro. DNA analysis for the sickle cell mutation is done by specific amplification of the DNA region spanning the mutation using polymerase chain reaction followed by enzymatic cleavage of the amplified product. Sickle cell mutation abolishes a restriction endonuclease site (Dde I). Electrophoretic resolution of the fragment pattern reveals the presence or absence of the mutation. Clear genotyping of normal, carrier and homozygous DNA is achieved. Protocol For Sickle Cell DNA Genotyping Material SuppliedTwo lyophilized oligonucleotide primers SC2 and SC5 are supplied. Each tube contains 10 nmoles of the primer. The quantity supplied is sufficient for 400 regular 50m l PCR reaction. Reconstitution 1. Stock solution: Add 50m l sterile water to each tube containing the primer. The 10 nmoles of primer when dissolved in 50m l water will give a solution of 200 m Molar i.e. 200 pmoles/m l. 2. Primer Mix: Prepare a 10 pmoles/m l Primer Mix solution containing BOTH the primers. Example; add 180 m l water to a new tube. To this tube add 10 m l of EACH of the primer stock solution from #1 step above. Label this tube as SC Primer Mix 10 pmoles/m l |
A. PCR* reaction (see Appendix for Details) b globin primersSC5 SC2 PCR Profile Denaturation 94oC 30 sec. Annealing 58oC 30 sec. Elongation 72oC 1 min.
30 cycles, 7 min. 72oC extension, 4oC soak.
B. Restriction enzyme digestion (100 m
l reaction)
Precipitate after over night digestion, dissolve pellets in 5m
l 1 x loading buffer.
C. Electrophoresis
D. Results
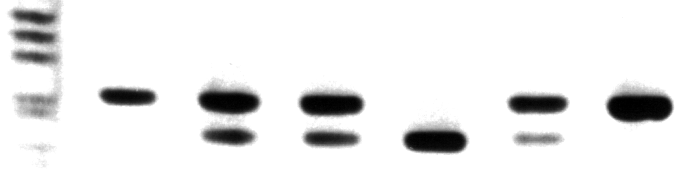
1 2 3 4 5 6 7 Figure 1. Typical Sickle cell genotype analysis of PCR product digested with Dde I. Lane 1 is molecular weight markers. Lane 2 is undigested PCR product. Lanes 3, 4 and 6 is DNA with A/S geneotype. Lane 5 is A/A genotype DNA and Lane 7 represents DNA with S/S genotype. References: 1. Saiki et al. (1985) Science 230:1350-1354 2. Wu et al. (1989) PNAS 86:2757-2760 3. Conner et al. (1983) PNAS 80:278-282 **The polymerase chain reaction (PCR) process is covered by patents owned by Hoffmann-La Roche. A license to perform is automatically granted by the use of authorized reagents.
All Gene Link products are for research use only. | |||||||||||||||
Appendix
| PCR
Premix preparation Typical Premix -----------------------------------------/50 >m l----rxn /1ml 10 x PCR Buffer ----------------___4.5 >m l___ 100>m l dNTP mix (2.5mM each) _________4 >m l___ 100>m l Primer Mix (10 pmol/>m l each) ___2.5 >m l____ 63>m l (25 pmol of each primer/50>m l) Sterile water _________________-34 >m l___ 737>m l ___________________________----------__-------- Total ________________________45 >m l____ 1ml Nucleotide
Dilution Total volume ________________________5.0ml |
Taq
Premix (per 50>m l reaction, scale up as
required) 10
x PCR Buffer ___________0.5>m l PCR reaction (50>m l) PCR products
post-processing |
Ordering Information
|
GENEMERä | |||
| Product | Size | Catalog No. | Price, $ |
|
Sickle Cell SC2/SC5 primer pair |
10nmoles |
40-2001-10 |
100.00 |
|
RhD (Rh D gene exon 10 specific) |
10nmoles |
40-2002-10 |
100.00 |
|
Rh EeCc (Rh Ee and Cc exon 7 specific) |
10nmoles |
40-2003-10 |
100.00 |
|
Fragile X (spanning triple repeat region) |
10nmoles |
40-2004-10 |
100.00 |
|
Gaucher 1226G mutation specific |
10nmoles |
40-2005-10 |
100.00 |
|
Gaucher 1448C mutation specific |
10nmoles |
40-2006-10 |
100.00 |
|
Gaucher 84GG mutation specific |
10nmoles |
40-2007-10 |
100.00 |
|
Gaucher IVS2 mutation specific |
10nmoles |
40-2008-10 |
100.00 |
|
Cystic Fibrosis D F508 |
10nmoles |
40-2009-10 |
100.00 |
|
Cystic Fibrosis G542X |
10nmoles |
40-2010-10 |
100.00 |
|
Cystic Fibrosis W1282X |
10nmoles |
40-2011-10 |
100.00 |
|
Cystic Fibrosis G551D/R553X |
10nmoles |
40-2012-10 |
100.00 |
|
Cystic Fibrosis N1303K |
10nmoles |
40-2013-10 |
100.00 |
|
Cystic FibrosisCT3849 |
10nmoles |
40-2014-10 |
100.00 |
|
SRY (sex determining region on Y) |
10nmoles |
40-2020-10 |
100.00 |
|
X alphoid repeat |
10nmoles |
40-2021-10 |
100.00 |
|
Y alphoid repeat |
10nmoles |
40-2022-10 |
100.00 |
LINKMERä
LINKMERä are primer pairs for specific amplification of chromosomal sequences; specially suited for linkage analysis and chromosomal mapping. LINKMERä are supplied as lyophilized powder in aliquots of 10nmoles. The 10nmoles of primer when dissolved in 500m l sterile water or TE will give a solution of 20m Molar i.e. 20pmoles/m l. The quantity supplied is sufficient for at least 400 regular 25m l PCR reaction.
| Product | Size | Catalog No. | Price |
|
Please inquire, Linkmersä |
10nmoles |
$75.00 |
**The polymerase chain reaction (PCR) process is covered by patents owned by Hoffmann-La Roche. A license to perform is automatically granted by the use of authorized reagents.
Prices subject to change without notice
All Gene Link products are for research use only.
Copyright Gene Link, Inc 2002. All Rights Reserved. For any help, please direct all inquiries to The Webmaster.