Technical Sheet
Rh D & EeCC
q Catalog No. 40-2002-10 Rh D 10 nmoles
q Catalog No. 40-2003-10 Rh CcEe 10 nmoles
Shipped at ambient temperature. Store at -20oC
For research use only.
The Rh based blood grouping is termed positive or negative based on the presence or absence of the D antigen. Rh alloimmunization in Rh negative pregnant women is of concern because of the potential for the fetus to develop hemolytic disease of newborns and autoimmune diseases. Anemia leading to hydrops, perinatal death, or both occurs in 25% of fetuses sensitized to the RhD antigen in the absence of optimal management. In utero diagnosis and treatment considerably improves the condition with survival rates greater than 75% in severely affected fetuses. However these invasive therapies may be unnecessary in some cases if the fetal Rh status were known prenatally.
A method of determining fetal Rh status early in pregnancy is now possible by DNA analysis of amniotic cells (1). The Rh blood group locus consists of two related structural genes, D and CcEe. These highly homologous genes which share greater than 96% identity in their coding region have been cloned and the molecular basis of the Rh blood group established (2). The RhD-positive and RhD-negative polymorphism is associated with the presence or the absence of the D gene (there is no ‘d’ gene). The C/c and E/e antigens are encoded by a unique gene. The E/e associated nucleotide polymorphism results in one amino acid substitution at position 226 (proline to alanine), whereas the C/c antigenic polymorphism consists of six nucleotide substitutions leading to four amino acid changes at position 16 (Cys16Trp), 60 (Ile60Leu), 68 (Ser68Asn) and 103 (Ser103Pro).
DNA analysis for Rh genotype loci specifically amplifies DNA fragments for the RhD and RhCcEe gene. Due to the high sensitivity and specificity of the test at the DNA level, occasionally the results may not match those obtained by serologic methods. The test will type individuals as RhD positive who are Du low grade status serologically (3-5), this is due to the absence of the gene product at the protein level due to partial deletions. The total error rate should be less than 1%.
Material Supplied
Two lyophilized oligonucleotide primers are supplied. Please refer to item number on the top of this sheet (Rh D or Rh EeCe Genemerä ). Each tube contains 10 nmoles of the primer. The quantity supplied is sufficient for 400 regular 50m l PCR reaction.
Stock solution
: Add 50m l sterile water to each tube containing the primer. The 10 nmoles of primer when dissolved in 50m l water will give a solution of 200 m Molar i.e. 200 pmoles/m l.Primer Mix: Prepare a 10 pmoles/m l Primer Mix solution containing BOTH the primers. Example; add 180 m l water to a new tube. To this tube add 10 m l of EACH of the primer stock solution from #1 step above. Label this tube as Primer Mix 10 pmoles/m l.
Protocol for RhD and Rh EeCc Genotyping
The following PCR* profile has been optimized for Rh D and Rh EeCc specific product amplification using the supplied Genemersä
PCR reaction (100 m l)
Denaturation 94oC 30 sec.
Annealing 49oC 1 min.
Elongation 72oC 1 min.
30 cycles; 7 min. at 72oC, soak at 4oC.
Precipitate products, dissolve in 10 m l water, add 2 m l loading buffer.
ElectrophoresisLoad samples in a 1.8 % agarose gel or a 3% Nusieve agarose gel. Run at 90 mAmps for 2.5 hrs.
Results and Interpretation
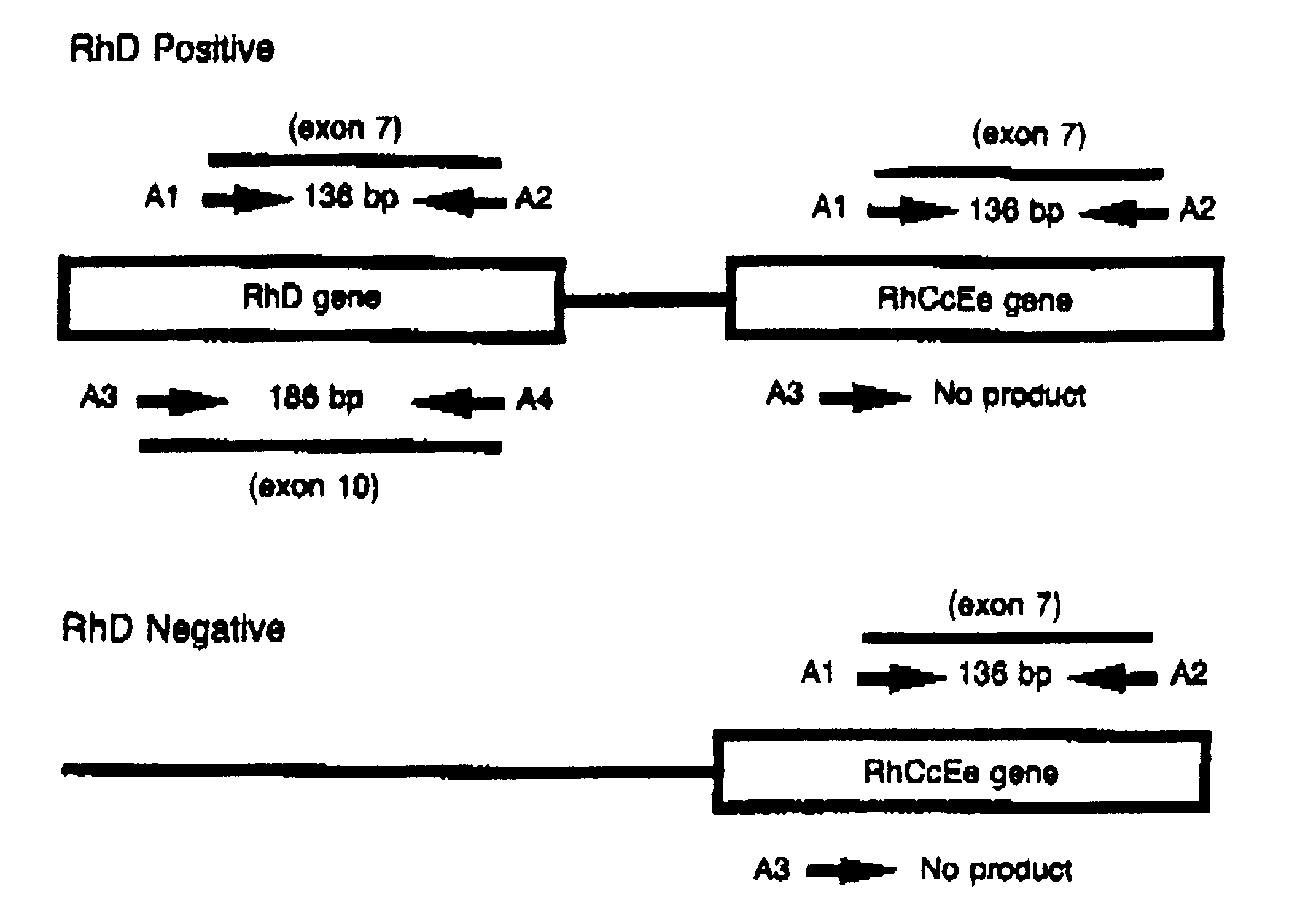
The primers A1/A2 will give 136 bp PCR product specific for Rh EeCc gene exon 7 (Catalog No. 40-2003-10) and primers A3/A4 will give 186 bp PCR product specific for Rh D gene exon 10 (Catalog No. 40-2002-10). Can perform multiplex of A1/A2 & A3/A4 as shown in the figure below
RhD negative- 136 bp only;
RhD positive- both 136 & 186 bp.
Figure. Lane 1 is molecular weight markers. Lanes 2-4 represent PCR from a Rh D positive DNA and Lanes 5-7 represent PCR from Rh D negative DNA. Lanes 2, 5 & 7 PCR product of A1/A2 amplification, lanes 3 from A3/A4 amplification. Lane 4 is a multiplex of A1/A2 and A3/A4 amplification. 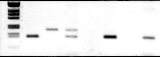
1 2 3 4 B 5 6 7
1. Bennet, P.R., et al. (1993) Prenatal determination of fetal RhD type by DNA amplification. NEJM 329:607-610.
2. Mouro, I., et al. (1993) Molecular genetic basis of the human Rhesus blood group system. Nature Genetics 5:62-65.
3. Simsek, S., Bleeker, P.M., Borne, A.. E.G. (1994) Prenatal determination of fetal RhD type. NEJM 330:795.
4. Bennet, P., Warwick,R. and Carton, J-P. (1994) Prenatal determination of fetal RhD type. NEJM 330:795-796.
Westhoff,C.M. and Wylie, D.E. (1994) Identification of a new RhD-specific mRNA from K562 cells. Blood 84:3098-3100