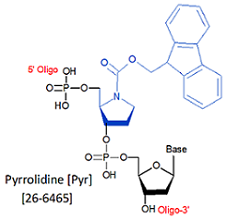
Modification : Pyrrolidine (Pyr)
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6465
Structural Studies
[Pyr]
Y
Y
Y
178.1
-
PS26-6465.pdf
-
-
-
| Catalog No | Scale | Price |
| 26-6465-05 | 50 nmol | $335.00 |
| 26-6465-02 | 200 nmol | $335.00 |
| 26-6465-01 | 1 umol | $395.00 |
| 26-6465-03 | 2 umol | $471.00 |
| 26-6465-06 | 5 umol | $1,777.50 |
| 26-6465-10 | 10 umol | $1,452.00 |
| 26-6465-15 | 15 umol | $1,884.00 |
| Discounts are available for Pyrrolidine (Pyr)! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Pyrrolidine: DNA Damage & Repair new base excision repair inhibitor
DNA is constantly under attack. Alkylating agents, ionizing radiation and oxidative stress can induce base modification or strand scission that, left unchecked, can lead to the development of cancer. Our cells are equipped with DNA damage-monitoring and repair enzymes to correct the damage via base excision repair (BER) or nucleotide excision repair (NER). Ironically, though, the up-regulation of these repair enzymes in cancerous cells frequently causes the development of drug resistance against chemotherapeutic reagents.
One of the most studied repair mechanisms is probably the base excision DNA repair pathway. In this pathway, DNA glycosylases recognize the damaged bases and catalyze their excision through hydrolysis of the N-glycosidic bond. Attempts to understand the structural basis for DNA damage recognition by DNA glycosylases have been hampered by the short-lived association of these enzymes with their DNA substrates. To overcome this problem, the design and synthesis of inhibitors that form stable complexes with DNA glycosylases are essential. Complexes can then be studied biochemically and structurally.
Toward this end, the Verdine group at Harvard synthesized a pyrrolidine analog that mimics the charged transition state of the enzyme-substrate complex. When incorporated into double-stranded DNA, they found the pyrrolidine analog (PYR), introduced through a synthetic oligo forms an extremely stable complex with the DNA glycosylase AlkA, exhibiting a dissociation constant in the pM range and potently inhibited the reaction catalyzed by the enzyme (1).
References
See Glen Research Report
DNA Damage & Repair.
1. O.D. Scharer, et al., J. Am. Chem. Soc., 1995, 117, 6623-6624.
2. O.D. Scharer, et al., J. Biol. Chem., 1998, 273, 8592-8597.
3. V.L. Schramm, Arch Biochem Biophys, 2005, 433, 13-26.
- Pyrrolidine (Pyr)
