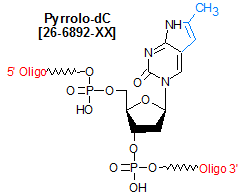
Modification : Pyrrolo-dC
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6892
Structural Studies
[Pyr-dC]
Y
Y
Y
327.23
2.41
PS26-6892.pdf
350
450
-
| Catalog No | Scale | Price |
| 26-6892-05 | 50 nmol | $346.00 |
| 26-6892-02 | 200 nmol | $346.00 |
| 26-6892-01 | 1 umol | $518.00 |
| 26-6892-03 | 2 umol | $622.00 |
| 26-6892-06 | 5 umol | $2,331.00 |
| 26-6892-10 | 10 umol | $4,147.00 |
| 26-6892-15 | 15 umol | $4,977.00 |
| Discounts are available for Pyrrolo-dC! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Pyrrolo-dC is a fluorescent bicyclic analog of deoxycytidine that base pairs specifically with deoxyguanosine (but not A, C, or T). Its excitation and emission maxima are 350 nm and 450 nm, respectively. Because these values are significantly different from the UV absorption maxima of DNA (260 nm) and protein (280 nm), pyrrolo-dC is useful for probing the structures of protein-nucleic acid complexes. This modification also shows reduced fluorescence in duplex DNA compared to in single-stranded DNA, a property that can be used to monitor local DNA melting behavior. These properties were particularly useful in studies done to characterize the nature and size of the transcription bubble in T7 RNA polymerase elongation complexes (1), the structure and function of the polypurine tract in HIV-1 (2) and the binding interactions between DNA hairpins and the anti-tumor drug actinomycin D (3). An important feature of pyrrolo-dC is that its fluorescence allows it to report directly on its local environment with minimal perturbation of the system.
Pyrrolo-dC modified oligonucleotides have also been used as part of signal-on aptasensor systems for molecular recognition. In one such system, an unmodified aptamer specific for a target functions as the molecular recognition element, and a pyrrolo-dC oligo functions as the signal transduction element. The latter acts as a signaling probe that responds to structural changes in aptamers that occur when they bind to targets (4).
References
1. Liu, C., Martin, C.T. Fluorescence Characterization of the Transcription Bubble in Elongation Complexes of T7 RNA Polymerase.
J. Mol. Biol. (2001),
308: 465-475.
2. Dash, C., Rausch, J.W., Le Grice, S.F.J. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3’ polypurine tract.
Nucleic Acids Res. (2004),
32: 1539-1547.
3. Xhang, X., Wadkins, R.M. DNA hairpins containing the cytidine analog pyrrolo-dC: structural, thermodynamic, and spectroscopic studies.
Biophys. J. (2009),
96: 21884-1891.
4. Li, T., Fu, R., Park, H.G. Pyrrolo-dC based fluorescent aptasensors for the molecular recognition of targets.
Chem. Commun. (2010),
46: 3271-3273.
- Pyrrolo-dC
