.png)
Modification : iso dC
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6992
Conjugation Chemistry
[iso-dC]
Y
Y
Y
289.19
7.4
PS26-6992.pdf
1
1
1
| Catalog No | Scale | Price |
| 26-6992-05 | 50 nmol | $379.00 |
| 26-6992-02 | 200 nmol | $379.00 |
| 26-6992-01 | 1 umol | $493.00 |
| 26-6992-03 | 2 umol | $739.00 |
| 26-6992-06 | 5 umol | $2,218.50 |
| 26-6992-10 | 10 umol | $3,942.00 |
| 26-6992-15 | 15 umol | $4,928.00 |
| Discounts are available for iso dC! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
iso dC and 5-methyl iso-deoxycytosine (5-Me-iso-dC) forms a Watson-Crick base pair with iso-dG, but has a different type of hydrogen bonding pattern than those observed for the natural base pairs A:T and C:G. Substitution of a 5-me iso-dC:iso-dG base pair for a C:G pair increases the Tm of the resulting duplex by ~2degC per base pair substitution (1,2). Furthermore, since iso dC and 5-Me-iso-dC does not pair with dG, iso dC and 5-Me-iso-dC:iso-dG can function as a stable unnatural base pair that can be used to expand the genetic code.
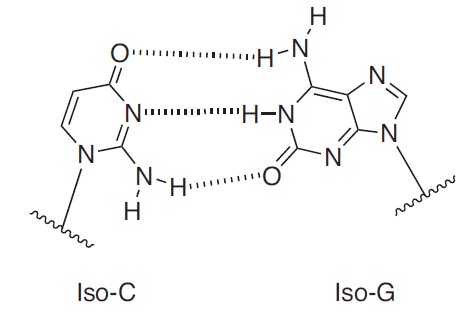
The combination of 5-Me-iso-dC’s high selectivity for iso-dG, and the resulting base pair’s high thermodynamic stability, make this modified base particular attractive in the following applications:
(a) Molecular recognition: The 5-Me-iso-dC:iso-dG base pair has been incorporated into hybridization assays to enhance probe-target specificity and reduce spurious hybridization to non-target sequences. For example, Collins and co-workers significantly improved the sensitivity of a branched DNA quantitative hybridization assay for detecting the HIV POL sequence by incorporating ~30% 5-Me-iso-dC and iso-dG into the pre-amplifier, branched DNA (bDNA) amplifier and alkaline phosphate probe sequences used in the assay (3). Use of this strategy resulted in a significant reduction in non-specific hybridization of the above three sequence types to non-target nucleic acid sequences, and thus less amplification of background. The limits of detection of the assay were improved 10-fold, from < 500 HIV molecules/mL to < 50 molecules/mL.
(b) qPCR and artificially expanded genetic systems: A number of research groups have been working on optimizing PCR amplification on templates containing 5-Me-iso-dC. Such optimization is necessary to enable the full development of artificially expanded genetic systems utilizing an expanded genetic code, thereby allowing for the site-specific incorporation of novel functional components (such as unnatural amino acids) into proteins. In 2004, Johnson and co-workers observed that, by using the Klenow fragment of Taq polymerase (KF-Taq) in PCR, the fidelity of the 5-Me-iso-dC:iso-dG base pair was about 96% per amplification cycle (4). The limit in fidelity is chiefly due to the ability of iso-dG’s 1,2 tautomer to mis-pair with dT. More recently, Sismour and Benner solved this problem by using 2-thio-dT (dT*) in place of dT. dT*pairs with dA, but not with iso-dG (5). Using this artificial base pair system (5-Me-iso-dC:iso-dG, dA:dT*, dC:dG) with KF-Taq, the fidelity in PCR was increased to about 98% per amplification cycle. The achievement of high fidelity PCR with the 5-Me-iso-dC:iso-dG base pair opens the door to developing both artificially expanded genetic systems (6) and novel qPCR systems (for example, Promega’s Plexor technology) based on this approach.
References
1. Switzer, C.; Moroney, S.E.; Benner, S.A. Enzymatic incorporation of a new base pair into DNA and RNA.
J. Am Chem. Soc. (1989),
111: 8322-8323.
2. Horn, T.; Chang C-A.; Collins, M.L. Hybridization properties of the 5-methy-isocytidine/isoguanosine base pair in synthetic oligodeoxynucleotides.
Tetrahedron Lett. (1995),
36: 2033-2036.
3. Collins, M.L.; Irvine, B.; Tyner, D.; Fine, E.; et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml.
Nucleic Acids Res. (1997),
25: 2979-2984.
4. Johnson, S.C.; Sherrill, C.B.; Marshall, D.J.; Moser, M.J.; Prudent, J.R. A third base pair for the polymerase chain reaction: inserting isoC and isoG.
Nucleic Acids Resl. (2004),
32: 1937-1941.
5. Sismour, A.M.; Benner, S.A. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system.
Nucleic Acids Resl. (2005),
33: 5640-5646.
6. Yang, Z.; Hutter, D.; Sheng, P.; Sismour, A.M.; Benner, S.A. Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern,
Nucleic Acids Res. (2006),
34: 6095-6101.
- iso deoxycytosine (iso dC)
