|
Technical Sheet
Non-radioactive southern detection of fragile X genotype
| Catalog Number: 40-3202-01 110m l Shipped at ambient temperature. Store at -20oC
For research use only.
Not
for use in diagnostic procedures for clinical
purposes. |
|
Background
Fragile X syndrome is the most common form of inherited
mental retardation. It affects approximately 1 in 1200 males and 1 in 2500 females. As
suggested by the name, it is associated with a fragile site under specific cytogenetic
laboratory conditions at position Xq27.3 (1).
The inheritance pattern of fragile X puzzled geneticists as it did not follow a clear X
linked pattern. Approximately 20% of males who are carriers based on pedigree analysis do
not manifest any clinical symptoms and are thus termed as Normal Transmitting Males (NTM),
mental retardation is rare among the daughters of male carriers. Approximately 35% of
female carriers have some mental impairment. Based on the above it has been proposed that
there are two states of the mutation, one mutation range in which there is no clinical
expression (premutation), which could change to the disease causing state predominantly
when transmitted by a female (full mutation)(2).
The fragile X syndrome gene (FMR-1, fragile X mental retardation -1) was cloned in 1991
simultaneously by three groups (3-6). Soon the peculiar genetic mode of transmission was
established and a new class of mutation came into existence- Triple repeat amplification.
This explained the clinical state of ‘premutation’ and ‘full mutation’
as well as ‘anticipation’. The fragile X syndrome is caused by the amplification
of CGG repeat which is located in the 5’ region of the cDNA. The most common allele
in the normal population consists of 29 repeats, the range varying from 6 to 54 repeats.
Premutations in fragile X families showing no phenotypic effect range in size from 52 to
over 200 repeats. All alleles with greater than 52 repeats are meiotically unstable with a
mutation frequency of one. In general repeats up to 45 are considered normal, repeats
above 50 to 200 are considered as premutation and above 200 as full mutation (3-7). The
range between 40-55 is considered even by most experienced clinical geneticists and
molecular geneticists very difficult to interpret and is considered as a ‘gray
zone’ with interpretations made on a case by case basis (8).
Genotyping
Fragile X genotyping can be done by direct PCR amplification of
the CGG triple repeat region or by southern analysis. In most cases both methods are used
to complement the results, full mutations usually cannot be identified by PCR by most
investigators and southern analysis is the preferred method to distinguish full mutations.
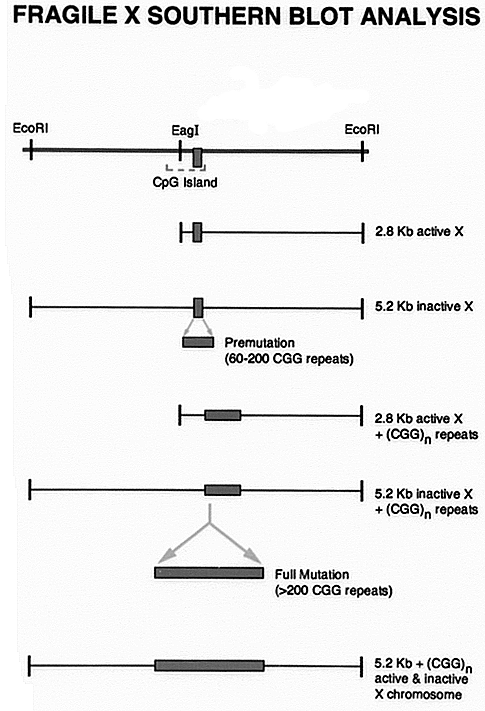
The FMR-1 gene region containing the CGG triple repeat is flanked by Eco RI sites and a
Eag I site in the region. Full mutation has been shown to methylate the active gene too
and thus it prevents Eag I restriction of DNA. Hybridization of southern blots of Eco RI
and Eag I double digested DNA clearly can distinguish between normal, premutation and full
mutation genotypes (2).
Southern analysis can not determine the exact number of repeats
or the identification of genotypes corresponding to the ‘gray zone’. |
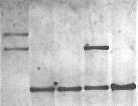
Fragile X southern blot. Lane 1
affected female.
Non-radioactive detection,
~2hr. exposure.
References
- Nelson, D.L. (1993) Growth Genetics and
Hormone. 9:1-4.
- Rousseau, F. et al. (1991) NEJM
325:1673-1681.
- Verkerk, A. et al. (1991) Cell 65:905-914
- Fu, Y.H et al. (1991) Cell 67:1047-1058.
- Oberle, I. et al. (1991) Science
252:1097-1102.
- Yu, S. et al. (1991) Science 252:
1179-1181.
- Nelson, D.L. (1996) Growth Gen. and
Hormone. 12:1-4.
- Richards, R and Sutherland, G.R (1992) TIG 8: 249-255.
|
**The polymerase chain reaction (PCR) process is covered by patents
owned by Hoffmann-La Roche.
A license to perform is automatically granted by the use of authorized
reagents.
All Gene Link products are for research use only.
Appendix GeneProberä GLFXDig1 40-3202-01
Material Supplied
One tube containing 110m l of GeneProberä GLFXDIG1 probe. This probe is digoxigenin labeled for non
radioactive detection. The quantity supplied is sufficient for at least 5 20x20 cm blots
using 20m l for each blot as probe.
Fragile X Southern Protocol
A. Chromosomal DNA digestion
DNA 10 mg
10x Eag I Buffer 10 ml
10x EcoR I Buffer 10 ml
Eag I (10 U/ ml) 4 ml
EcoR I( 10 U/ ml) 8 ml
------------------------
H2O up to 100 ml
Precipitate the digests after over night digestion at 37oC, dissolve the pellets in 10 ml of 1x Loading
buffer .
B. Electrophoresis and Transfer
1. Load samples to 0.8% agarose gel , run over night at 45mA for 20-24 hours. (1.6
kb fragment on the bottom of the gel).
2. Depurinate with 0.25N HCl (add 10 ml HCl to 500ml H2O) for 10 minutes,
denature the DNA with 0.4N NaOH/0.6M NaCl for 30 min. at RT, neutralize with 1.5M
NaCl/0.5M Tris ( pH 7.5) for 30 min. at RT, transfer to the BM positively charged nylon
membrane using 10xSSC and 10 pieces of SIGMA QuickDraw blotting paper overnight. Wash the
membrane with 2x SSC, bake at 80oC for 2 hours. |
C. Hybridization
1. Perform prehybridization at 55oC for 3 hours in 10 ml of Easy Hyb buffer
(Boehringer Mannheim) .
2. Boil 20m l GeneProberä GLFXDIG1
probe in 500m l of Easy Hyb for 10 minutes. Chill directly on
ice. Add the above probe to10ml of Easy Hyb. Discard the prehybridization buffer and
replace it with the hybridization buffer containing the boiled probe. Hybridize overnight
at 55oC.
Gene Link recommends using Boehringer Mannheim Digoxigenin based
washing and detection system.
3. Detection with CDP star(Tropix) as substrate will yield reliable result by
exposing X-ray film for 1 hour to overnight at room temperature.
D. Stripping
Wash the membrane in water to remove the substrate. Then wash the membrane in 0.2N
NaOH/0.1% SDS at 37oC for 30 minutes. Rinse the membrane in 2XSSC. Air dry.
Call Gene Link Technical service for more information.
1-800-Gene Link (1-800-436-3546) |
Ordering
Information
| GeneProberä |
| Product |
Size |
Catalog No. |
Price, $ |
| Fragile X GLFX1 Suitable for random primer
labeling |
500ng |
40-3201-01 (old number 40-2015-10) |
350.00 |
| Fragile X GLFXDig1 Digoxigenin labeled
probe, ready to use for southern hybridization |
110m l |
40-3202-01 |
400.00 |
| Fragile X PCR Probe; GLFXPCRprober For non
radioactive detection of Fragile X PCR products |
5blots |
40-3101-01 |
400.00 |
| |
| GENEMERä |
| Product |
Size |
Catalog No. |
Price, $ |
| Sickle Cell SC2/SC5 primer pair |
10nmoles |
40-2001-10 |
100.00 |
| RhD (Rh D gene exon 10 specific) |
10nmoles |
40-2002-10 |
100.00 |
| Rh EeCc (Rh Ee and Cc exon 7 specific) |
10nmoles |
40-2003-10 |
100.00 |
| Fragile X (spanning triple repeat region) |
10nmoles |
40-2004-10 |
100.00 |
| Gaucher 1226G mutation specific |
10nmoles |
40-2005-10 |
100.00 |
| Gaucher 1448C mutation specific |
10nmoles |
40-2006-10 |
100.00 |
| Gaucher 84GG mutation specific |
10nmoles |
40-2007-10 |
100.00 |
| Gaucher IVS2 mutation specific |
10nmoles |
40-2008-10 |
100.00 |
| Cystic Fibrosis D
F508 |
10nmoles |
40-2009-10 |
100.00 |
| Cystic Fibrosis G542X |
10nmoles |
40-2010-10 |
100.00 |
| Cystic Fibrosis W1282X |
10nmoles |
40-2011-10 |
100.00 |
| Cystic Fibrosis G551D/R553X |
10nmoles |
40-2012-10 |
100.00 |
| Cystic Fibrosis N1303K |
10nmoles |
40-2013-10 |
100.00 |
| Cystic FibrosisCT3849 |
10nmoles |
40-2014-10 |
100.00 |
| SRY (sex determining region on Y) |
10nmoles |
40-2020-10 |
100.00 |
| X alphoid repeat |
10nmoles |
40-2021-10 |
100.00 |
| Y alphoid repeat |
10nmoles |
40-2022-10 |
100.00 |
| |
|
|
|
| |
|
|
|
Please inquire about
other GENEMERä not listed here |
Revised 10/27/1997
**The polymerase chain reaction (PCR) process is covered by patents
owned by Hoffmann-La Roche. A license to perform is automatically granted by the use of
authorized reagents.
**The polymerase chain reaction (PCR) process is covered by patents
owned by
Hoffmann-La Roche. A license to perform is automatically granted by the
use of authorized reagents.
Prices subject to change without notice
All Gene Link products are for research use only.
Home |
Products | Order |
Literature | Contact Us|
Site Map
Copyright Gene Link, Inc 2002. All Rights Reserved. For any help, please direct all inquiries to The Webmaster. |


