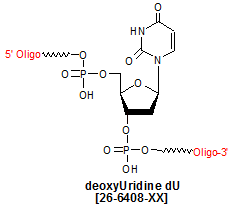
Modification : dU
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6408
DNA Oligo Synthesis
[dU]
Y
Y
Y
290.17
8.4
PS26-6408.pdf
-
-
-
| Catalog No | Scale | Price |
| 26-6408-05 | 50 nmol | $27.00 |
| 26-6408-02 | 200 nmol | $27.00 |
| 26-6408-01 | 1 umol | $59.00 |
| 26-6408-03 | 2 umol | $119.00 |
| 26-6408-06 | 5 umol | $265.50 |
| 26-6408-10 | 10 umol | $475.00 |
| 26-6408-15 | 15 umol | $594.00 |
| Discounts are available for dU! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Deoxyuridine (dU) is a pyrimidine deoxyribonucleoside, and a derivative of the nucleoside uridine, with the only difference being that, in dU, a hydrogen (-H) group is substituted for uridine�s �OH group located at the 2�-position of the ribose. dU is generated in cellular DNA as a deamination product of dC (deoxycytidine), with the deamination process catalyzed by the enzyme AID (activation-induced cytidine deaminase) (1). AID is a B cell-specific gene that is necessary for antibody gene diversification via class-switch recombination and somatic hypermutation (2, 3). The dC-to-dU conversion(s) by AID occurs in the IgG locus, with various gene diversification pathways arising from the different DNA repair mechanisms used by B-cells to repair the dU lesion (1).
dC-to-dU conversion via cytidine deamination is also implicated in innate immunity to retroviruses. Here deamination of dC is mediated by the enzyme APOBEC3G, which is present in T cells, acting on the first (minus) strand cDNA of retroviruses. Generation of dU produces a dU /dG mismatch in the retroviral cDNA duplex, resulting in a dC-to-dT transition mutation on the minus-strand cDNA, and a dG-to-dA transition on the plus-strand (4). The presence of dU in the minus-strand cDNA could lead to innate immunity by one or more of the following: (a) hypermutation capable of disabling viral functions, (b) degradation by BER (base excision repair), (c) plus-strand cDNA mis-replication (5). dU can be used to modify oligos for use in studies of DNA damage and associated repair mechanisms.
Oligos modified with dU can serve as effective research tools for mechanistic studies of both adaptive and innate immunity in animal systems.
1. Neuberger, M.S., Harris, R.S., Di Noia, J., Petersen-Mahrt, S.K. Immunity through DNA deamination.
Trends Biochem. Sci. (2003),
28: 305-312.
2. Muramatsu, M., Kinoshita, K., Fagarason, S., Yamada, S., Shinkai, Y., Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme.
Cell (2000),
102: 553-563.
3. Revy, P., Muto, T., Levy, Y., Geissman, F., et al. Activation-Induced Cytidine Deaminase (AID) Deficiency Causes the Autosomal Recessive Form of the Hyper-IgM Syndrome (HIGM2).
Cell (2000),
102: 565-575.
4. Lecossier, D., Bouchonnet, F., Clavel, F., Hance, A.J. Hypermutation of HIV-1 DNA in the Absence of the Vif Protein.
Science (2003),
300: 1112.
5. Harris, R.S., Sheehy, A.M., Craig, H.M., Malim, M.H., Neuberger, M.S. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses.
Nature Immunology (2003),
4: 641-643.
- deoxyuridine dU
