In this section: Introduction | Quality Control | Purification | Modifications | Long Oligos | Price List
In this section: Introduction | Molecular Beacon FAQ's | Fluorescent Probes Price List | Other Fluorescent Molecular Probes
In this section: SPCT | DME SPCT Intro | Order DME TaqMan® Assays SPCT | SNP PCT Search | Gene Expression Assays | SPCT Design Center | GeneAssays
In this section: RNA Oligonucleotides | Quality Control | Purification | Modifications | RNAi Explorer™ Products and Prices | Custom RNAi | RNAi Design Guidelines | SmartBase™ siRNA Modifications | shRNA Explorer™
In this section: PCR Amplification & Analysis
In this section: Introduction | Genemer™ | GeneProber™ | Prober™ Gene Detection Kits | GScan™ Gene Detection Kits | Genemer™ Control DNA | Infectious Diseases
In this section: Gene Construction
In this section: Introduction | The Omni-Clean™ System | The Omni-Pure™ Plasmid Purification System | The Omni-Pure™ Genomic DNA Purification System | Viral DNA & RNA Purification | Microbial DNA Purification | Plant DNA Purification
In this section: Introduction | Quality Control | Purification | Modifications | Long Oligos | Price List
In this section: Introduction | Molecular Beacon FAQ's | Fluorescent Probes Price List | Other Fluorescent Molecular Probes
In this section: SPCT | DME SPCT Intro | Order DME TaqMan® Assays SPCT | SNP PCT Search | Gene Expression Assays | SPCT Design Center | GeneAssays
In this section: RNA Oligonucleotides | Quality Control | Purification | Modifications | RNAi Explorer™ Products and Prices | Custom RNAi | RNAi Design Guidelines | SmartBase™ siRNA Modifications | shRNA Explorer™
In this section: PCR Amplification & Analysis
In this section: Introduction | Genemer™ | GeneProber™ | Prober™ Gene Detection Kits | GScan™ Gene Detection Kits | Genemer™ Control DNA | Infectious Diseases
In this section: Gene Construction
In this section: Introduction | The Omni-Clean™ System | The Omni-Pure™ Plasmid Purification System | The Omni-Pure™ Genomic DNA Purification System | Viral DNA & RNA Purification | Microbial DNA Purification | Plant DNA Purification
Locked Analog A (LNA-A)
Locked Analog A
Code : [+A]
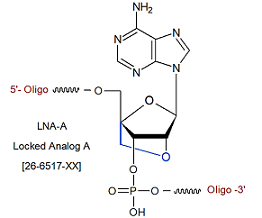
Modification : Locked Analog A
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6517
Duplex Stability
[+A]
Y
Y
Y
341.21
15.4
PS26-6517.pdf
260
-
-
| Catalog No | Scale | Price | | 26-6517-05 | 50 nmol | $41.00 | | 26-6517-02 | 200 nmol | $41.00 | | 26-6517-01 | 1 umol | $49.00 | | 26-6517-03 | 2 umol | $81.00 | | 26-6517-06 | 5 umol | $220.50 | | 26-6517-10 | 10 umol | $340.00 | | 26-6517-15 | 15 umol | $443.00 |
| Discounts are available for Locked Analog A! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
Antisense Oligos (ODN) & siRNA Oligo Modifications
Click here for more information on antisense modifications, design & applications.
General Locked Nucleic Acid Analog Oligonucleotide Design Guidelines
1. Locked Nucleic Acid Analog (LNA) should be introduced at the positions where specificity and discrimination is needed (e.g. 3'-end in allele specific PCR and in the SNP position in allele specific hybridization probes).
2. Avoid stretches of more than 4 LNA bases. LNA hybridizes very tightly when several consecutive residues are substituted with Locked Nucleic Acid Analog bases.
3. Avoid LNA self-complementarity and complementarity to other LNA containing oligonucleotides in the assay. LNA binds very tightly to other LNA residues.
4. Typical primer length of 18mer should not contain more than 6-8 LNA bases.
5. Each LNA bases increases the Tm by approximately 2-4oC.
6. Do not use blocks of LNA near the 3'-end.
7. Keep the GC-content between 30-60 %.
8. Avoid stretches of more than 3 G DNA or LNA bases.
9. Tm of the primer pairs should be nearly equal.
LNA is a bicyclic nucleic acid where a ribonucleoside is linked between the 2'-oxygen and the 4'-carbon atoms with a methylene unit. Locked Nucleic Acid (LNA) was first described by Wengel and co-workers in 1998 (8-10) as a novel class of conformationally restricted oligonucleotide analogues.
The design and ability of oligos containing locked nucleic acids (LNAs) to bind supercoiled, double-stranded plasmid DNA in a sequence-specific manner has been described by Hertoghs et al (6) for the first time. The main mechanism for LNA oligos binding plasmid DNA is demonstrated to be by strand displacement. LNA oligos are more stably bound to plasmid DNA than similar peptide nucleic acid (PNA) `clamps' for procedures such as particle mediated DNA delivery (gene gun). It is shown that LNA oligos remain associated with plasmid DNA after cationic lipid-mediated transfection into mammalian cells. LNA oligos can bind to DNA in a sequence-specific manner so that binding does not interfere with plasmid conformation or gene expression (6).
LNA Oligonucleotides exhibit unprecedented thermal stabilities towards complementary DNA and RNA, which allow excellent mismatch discrimination (8). The high binding affinity of LNA oligos allows for the use of short probes in antisense protocols and LNA is recommended for use in any hybridization assay that requires high specificity and/or reproducibility, e.g., dual labeled probes, in situ hybridization probes, molecular beacons and PCR primers. Furthermore, LNA offers the possibility to adjust Tm values of primers and probes in multiplex assays. Each LNA base addition in an oligo increases the Tm by approximately 8oC. As a result of these significant characteristics, the use of LNA-modified oligos in antisense drug development is now coming under investigation, and recently the therapeutic potential of LNA has been reviewed (11).
Modifications Increasing Duplex Stability and Nuclease Resistance
|
Modification
|
Duplex Stability [Tm Increase]
|
Nuclease Resistance
|
| Locked Analog Bases |
Increased [2- 4C per substitution] |
Increased |
| 2-Amino-dA |
Increased [3.0C per substitution] |
Similar to DNA |
| C-5 propynyl-C |
Increased [2.8C per substitution] |
Increased |
| C-5 propynyl-U |
Increased [1.7C per substitution] |
Increased |
| 2'-Fluoro |
Increased [1.8C per substitution] |
Increased |
| 5-Methyl-dC |
Increased [1.3C per substitution] |
Similar to DNA |
| 2'-O Methyl |
Increased |
Increased |
| Phosphorothioate |
Slightly decreased |
Increased |
|
Click here for complete list of duplex stability modifications
|
The synthesis and incorporation of LNA bases can be achieved by using standard DNA synthesis chemistry. Detailed research results have not yet concluded as to the amount of LNA bases and regular DNA base combination in successful antisense and gene delivery experiments. The investigator can elect to substitute individual bases in the oligo to LNA bases or use a combination. Due to the high affinity and thermal stability of the LNA: DNA duplex it is not advised to have more than 15 LNA bases in an oligo; this induces strong self-hybridization The use of LNA C base requires special synthesis and post synthesis protocols. LNA-containing oligonucleotides can be purified and analyzed using the same methods employed for standard DNA. LNA can be mixed with DNA and RNA, as well as other nucleic acid analogues, modifiers and labels. LNA oligonucleotides are water soluble, and can be separated by gel electrophoresis and precipitated by ethanol.
- Locked Analog A (LNA-A)
|
|
