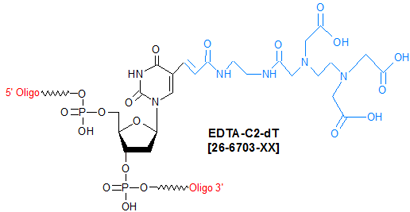
Modification : EDTA-C2-dT
Catalog Reference Number
Category
Modification Code
5 Prime
3 Prime
Internal
Molecular Weight (mw)
Extinction Coeficient (ec)
Technical Info (pdf)
Absorbance MAX
Emission MAX
Absorbance EC
26-6703
Minor Bases
[EDTA-C2-dT]
Y
Y
Y
676.53
8.7
PS26-6703.pdf
-
-
-
| Catalog No | Scale | Price |
| 26-6703-05 | 50 nmol | $378.00 |
| 26-6703-02 | 200 nmol | $378.00 |
| 26-6703-01 | 1 umol | $491.00 |
| 26-6703-03 | 2 umol | $737.00 |
| 26-6703-06 | 5 umol | $2,209.50 |
| 26-6703-10 | 10 umol | $3,931.00 |
| 26-6703-15 | 15 umol | $4,914.00 |
| Discounts are available for EDTA-C2-dT! |
| Modification* Discount Price Structure |
|
1 site/order
|
List price
|
|
2 sites/order
|
10% discount
|
|
3 sites/order
|
20% discount
|
|
4 sites/order
|
30% discount
|
|
5-9 sites/order
|
50% discount
|
|
10+ sites/order
|
60% discount
|
|
*Exceptions apply
|
EDTA-C2-dT is a deoxythymidine conjugated with the triethanoic (that is, triacetic) acid derivative of EDTA at the C-5 position of the thymine base. Oligonucleotides containing EDTA-C2-dT are typically used as artificial nucleases for cleaving both single and double-stranded DNA (1,2). In such cases, the modified oligo serves as a molecular recognition moiety, while the EDTA complexed to a metal ion (such as Fe(II)) acts as the actual cleavage agent. This analytical approach has been used to investigate the tertiary structure of tRNA (3).
Note that, because addition of Fe(II) and dithiothreotol (DTT) to form the Fe-EDTA complex in the oligo results in fairly rapid autocleavage of the oligo’s own phosphodiester backbone (thereby rendering the oligo useless as an artificial nuclease), substitution of methylphosphonate into this backbone can be done to confer resistance to such autocleavage (4).
Besides being used in the synthesis of oligo-based artificial nucleases, another possible use for EDTA-C2-dT is to introduce a metal complex (for example, a Cu-64-EDTA complex) into an oligonucleotide probe for use in experiments requiring in vivo imaging with positron emission tomography (PET).
References
1. Dreyer, G.B., Dervan, P.B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II).
Proc. Natl Acad. Sci. USA (1985),
82: 968-972.
2. Moser, H.E., Dervan, P.B. Sequence-specific cleavage of double helical DNA by triple helix formation.
Science (1987),
238: 645-650.
3. Han, H., Dervan, P.B. Visualization of RNA tertiary structure by RNA-EDTA X Fe(II) autocleavage: analysis of tRNA(Phe) with uridine-EDTA X Fe(II) at position 47.
Proc. Natl. Acad. Sci. USA. (1994),
91: 4955-4959.
4. Lin, S-B., Blake, K.R., Miller, P.S., Ts’o, P.O. Use of EDTA Derivitization To Characterize Interactions between Oligodeoxyribonucleoside Methylphosphonates and Nucleic Acids.
Biochemistry (1989),
28: 1054-1061.
- EDTA-C2-dT
